Proteomics Computational Analyses Suggest that the Antennavirus Glycoprotein Complex Includes a Class I Viral Fusion Protein (α-Penetrene) with an Internal Zinc-Binding Domain and a Stable Signal Peptide
Abstract
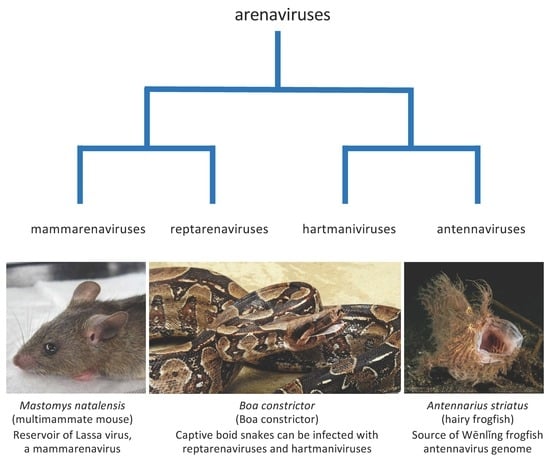
:1. Introduction
2. Materials and Methods
2.1. Sequences
2.2. Proteomics Computational Methods
3. Results
3.1. Antennavirus Glycoprotein 2 Has Features of a Class I Viral Fusion Protein
3.2. Reptarenavirus Glycoprotein 2 Is a Filovirus-Like Viral Fusion Protein
3.3. Antennavirus Glycoprotein 1 Does Not Display Features Conserved in Glycoprotein 1 of Other Arenaviruses
3.4. Antennavirus Nucleoprotein and Large Proteins Share Similarities to Hartmanivirus Nucleoprotein and Large Proteins
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Radoshitzky, S.R.; Bao, Y.; Buchmeier, M.J.; Charrel, R.N.; Clawson, A.N.; Clegg, C.S.; DeRisi, J.L.; Emonet, S.; Gonzalez, J.P.; Kuhn, J.H.; et al. Past, present, and future of arenavirus taxonomy. Arch. Virol. 2015, 160, 1851–1874. [Google Scholar] [CrossRef] [PubMed]
- Cajimat, M.N.; Milazzo, M.L.; Haynie, M.L.; Hanson, J.D.; Bradley, R.D.; Fulhorst, C.F. Diversity and phylogenetic relationships among the north american tacaribe serocomplex viruses (family arenaviridae). Virology 2011, 421, 87–95. [Google Scholar] [CrossRef] [PubMed]
- Charrel, R.N.; Feldmann, H.; Fulhorst, C.F.; Khelifa, R.; de Chesse, R.; de Lamballerie, X. Phylogeny of new world arenaviruses based on the complete coding sequences of the small genomic segment identified an evolutionary lineage produced by intrasegmental recombination. Biochem. Biophys. Res. Commun. 2002, 296, 1118–1124. [Google Scholar] [CrossRef]
- Downs, W.G.; Anderson, C.R.; Spence, L.; Aitken, T.H.; Greenhall, A.H. Tacaribe virus, a new agent isolated from artibeus bats and mosquitoes in trinidad, west indies. Am. J. Trop. Med. Hyg. 1963, 12, 640–646. [Google Scholar] [CrossRef] [PubMed]
- Milazzo, M.L.; Cajimat, M.N.; Mauldin, M.R.; Bennett, S.G.; Hess, B.D.; Rood, M.P.; Conlan, C.A.; Nguyen, K.; Wekesa, J.W.; Ramos, R.D.; et al. Epizootiology of tacaribe serocomplex viruses (arenaviridae) associated with neotomine rodents (cricetidae, neotominae) in southern california. Vector Borne Zoonotic Dis. 2015, 15, 156–166. [Google Scholar] [CrossRef] [PubMed]
- Fulhorst, C.F.; Bowen, M.D.; Salas, R.A.; Duno, G.; Utrera, A.; Ksiazek, T.G.; De Manzione, N.M.; De Miller, E.; Vasquez, C.; Peters, C.J.; et al. Natural rodent host associations of guanarito and pirital viruses (family arenaviridae) in central venezuela. Am. J. Trop. Med. Hyg. 1999, 61, 325–330. [Google Scholar] [CrossRef] [PubMed]
- Kronmann, K.C.; Nimo-Paintsil, S.; Guirguis, F.; Kronmann, L.C.; Bonney, K.; Obiri-Danso, K.; Ampofo, W.; Fichet-Calvet, E. Two novel arenaviruses detected in pygmy mice, ghana. Emerg. Infect. Dis. 2013, 19, 1832–1835. [Google Scholar] [CrossRef]
- Olayemi, A.; Cadar, D.; Magassouba, N.; Obadare, A.; Kourouma, F.; Oyeyiola, A.; Fasogbon, S.; Igbokwe, J.; Rieger, T.; Bockholt, S.; et al. New hosts of the lassa virus. Sci. Rep. 2016, 6, 25280. [Google Scholar] [CrossRef]
- Monath, T.P.; Newhouse, V.F.; Kemp, G.E.; Setzer, H.W.; Cacciapuoti, A. Lassa virus isolation from mastomys natalensis rodents during an epidemic in sierra leone. Science 1974, 185, 263–265. [Google Scholar] [CrossRef]
- Rowe, W.P.; Murphy, F.A.; Bergold, G.H.; Casals, J.; Hotchin, J.; Johnson, K.M.; Lehmann-Grube, F.; Mims, C.A.; Traub, E.; Webb, P.A. Arenoviruses: Proposed name for a newly defined virus group. J. Virol. 1970, 5, 651–652. [Google Scholar]
- Li, K.; Lin, X.D.; Li, M.H.; Wang, M.R.; Sun, X.Y.; Zhang, Y.Z. Genomic analysis of wenzhou virus in rodents from zhejiang province. Zhonghua Liu Xing Bing Xue Za Zhi 2017, 38, 384–387. [Google Scholar] [PubMed]
- Blasdell, K.R.; Becker, S.D.; Hurst, J.; Begon, M.; Bennett, M. Host range and genetic diversity of arenaviruses in rodents, united kingdom. Emerg. Infect. Dis. 2008, 14, 1455–1458. [Google Scholar] [CrossRef] [PubMed]
- Li, K.; Lin, X.D.; Wang, W.; Shi, M.; Guo, W.P.; Zhang, X.H.; Xing, J.G.; He, J.R.; Wang, K.; Li, M.H.; et al. Isolation and characterization of a novel arenavirus harbored by rodents and shrews in zhejiang province, China. Virology 2015, 476, 37–42. [Google Scholar] [CrossRef] [PubMed]
- Shaffer, J.G.; Grant, D.S.; Schieffelin, J.S.; Boisen, M.L.; Goba, A.; Hartnett, J.N.; Levy, D.C.; Yenni, R.E.; Moses, L.M.; Fullah, M.; et al. Lassa fever in post-conflict sierra leone. PLoS Negl. Trop. Dis. 2014, 8, e2748. [Google Scholar] [CrossRef] [PubMed]
- Okokhere, P.; Colubri, A.; Azubike, C.; Iruolagbe, C.; Osazuwa, O.; Tabrizi, S.; Chin, E.; Asad, S.; Ediale, E.; Rafiu, M.; et al. Clinical and laboratory predictors of lassa fever outcome in a dedicated treatment facility in nigeria: A retrospective, observational cohort study. Lancet. Infect. Dis. 2018, 18, 684–695. [Google Scholar] [CrossRef]
- Mehand, M.S.; Al-Shorbaji, F.; Millett, P.; Murgue, B. The who r&d blueprint: 2018 review of emerging infectious diseases requiring urgent research and development efforts. Antiviral Res. 2018, 159, 63–67. [Google Scholar]
- Burki, T. Cepi: Preparing for the worst. Lancet. Infect. Dis. 2017, 17, 265–266. [Google Scholar] [CrossRef]
- Stenglein, M.D.; Sanders, C.; Kistler, A.L.; Ruby, J.G.; Franco, J.Y.; Reavill, D.R.; Dunker, F.; Derisi, J.L. Identification, characterization, and in vitro culture of highly divergent arenaviruses from boa constrictors and annulated tree boas: Candidate etiological agents for snake inclusion body disease. MBio 2012, 3, e00180-12. [Google Scholar] [CrossRef]
- Abba, Y.; Hassim, H.; Hamzah, H.; Ibrahim, O.E.; Ilyasu, Y.; Bande, F.; Mohd Lila, M.A.; Noordin, M.M. In vitro isolation and molecular identification of reptarenavirus in malaysia. Virus Genes 2016, 52, 640–650. [Google Scholar] [CrossRef]
- Keller, S.; Hetzel, U.; Sironen, T.; Korzyukov, Y.; Vapalahti, O.; Kipar, A.; Hepojoki, J. Co-infecting reptarenaviruses can be vertically transmitted in boa constrictor. PLoS Pathog 2017, 13, e1006179. [Google Scholar] [CrossRef]
- Bodewes, R.; Kik, M.J.; Raj, V.S.; Schapendonk, C.M.; Haagmans, B.L.; Smits, S.L.; Osterhaus, A.D. Detection of novel divergent arenaviruses in boid snakes with inclusion body disease in the netherlands. J. Gen. Virol. 2013, 94, 1206–1210. [Google Scholar] [CrossRef] [PubMed]
- Bodewes, R.; Raj, V.S.; Kik, M.J.; Schapendonk, C.M.; Haagmans, B.L.; Smits, S.L.; Osterhaus, A.D. Updated phylogenetic analysis of arenaviruses detected in boid snakes. J. Virol. 2014, 88, 1399–1400. [Google Scholar] [CrossRef] [PubMed]
- Hetzel, U.; Sironen, T.; Laurinmaki, P.; Liljeroos, L.; Patjas, A.; Henttonen, H.; Vaheri, A.; Artelt, A.; Kipar, A.; Butcher, S.J.; et al. Isolation, identification, and characterization of novel arenaviruses, the etiological agents of boid inclusion body disease. J. Virol. 2013, 87, 10918–10935. [Google Scholar] [CrossRef] [PubMed]
- Hetzel, U.; Sironen, T.; Laurinmaki, P.; Liljeroos, L.; Patjas, A.; Henttonen, H.; Vaheri, A.; Artelt, A.; Kipar, A.; Butcher, S.J.; et al. Reply to updated phylogenetic analysis of arenaviruses detected in boid snakes. J. Virol. 2014, 88, 1401. [Google Scholar] [CrossRef] [PubMed]
- Hepojoki, J.; Hepojoki, S.; Smura, T.; Szirovicza, L.; Dervas, E.; Prahauser, B.; Nufer, L.; Schraner, E.M.; Vapalahti, O.; Kipar, A.; et al. Characterization of haartman institute snake virus-1 (hisv-1) and hisv-like viruses-the representatives of genus hartmanivirus, family arenaviridae. PLoS Pathog. 2018, 14, e1007415. [Google Scholar] [CrossRef] [PubMed]
- Hepojoki, J.; Salmenpera, P.; Sironen, T.; Hetzel, U.; Korzyukov, Y.; Kipar, A.; Vapalahti, O. Arenavirus coinfections are common in snakes with boid inclusion body disease. J. Virol. 2015, 89, 8657–8660. [Google Scholar] [CrossRef] [PubMed]
- Maes, P.; Adkins, S.; Alkhovsky, S.V.; Avsic-Zupanc, T.; Ballinger, M.J.; Bente, D.A.; Beer, M.; Bergeron, E.; Blair, C.D.; Briese, T.; et al. Taxonomy of the order bunyavirales: Second update 2018. Arch. Virol. 2019, 164, 927–941. [Google Scholar] [CrossRef] [PubMed]
- Maes, P.; Alkhovsky, S.V.; Bao, Y.; Beer, M.; Birkhead, M.; Briese, T.; Buchmeier, M.J.; Calisher, C.H.; Charrel, R.N.; Choi, I.R.; et al. Taxonomy of the family arenaviridae and the order bunyavirales: Update 2018. Arch. Virol. 2018, 163, 2295–2310. [Google Scholar] [CrossRef] [PubMed]
- Hyndman, T.H.; Marschang, R.E.; Bruce, M.; Clark, P.; Vitali, S.D. Reptarenaviruses in apparently healthy snakes in an australian zoological collection. Aust. Vet. J. 2019, 97, 93–102. [Google Scholar] [CrossRef]
- Stenglein, M.D.; Sanchez-Migallon Guzman, D.; Garcia, V.E.; Layton, M.L.; Hoon-Hanks, L.L.; Boback, S.M.; Keel, M.K.; Drazenovich, T.; Hawkins, M.G.; DeRisi, J.L. Differential disease susceptibilities in experimentally reptarenavirus-infected boa constrictors and ball pythons. J. Virol. 2017, 91. [Google Scholar] [CrossRef]
- Shi, M.; Lin, X.D.; Chen, X.; Tian, J.H.; Chen, L.J.; Li, K.; Wang, W.; Eden, J.S.; Shen, J.J.; Liu, L.; et al. The evolutionary history of vertebrate rna viruses. Nature 2018, 556, 197–202. [Google Scholar] [CrossRef] [PubMed]
- Abudurexiti, A.; Adkins, S.; Alioto, D.; Alkhovsky, S.V.; Avsic-Zupanc, T.; Ballinger, M.J.; Bente, D.A.; Beer, M.; Bergeron, E.; Blair, C.D.; et al. Taxonomy of the order bunyavirales: Update 2019. Arch. Virol. 2019, 164, 1949–1965. [Google Scholar] [CrossRef] [PubMed]
- Pontremoli, C.; Forni, D.; Sironi, M. Arenavirus genomics: Novel insights into viral diversity, origin, and evolution. Curr. Opin. Virol. 2019, 34, 18–28. [Google Scholar] [CrossRef] [PubMed]
- Hastie, K.M.; Zandonatti, M.; Liu, T.; Li, S.; Woods, V.L., Jr.; Saphire, E.O. Crystal structure of the oligomeric form of lassa virus matrix protein z. J. Virol. 2016, 90, 4556–4562. [Google Scholar] [CrossRef] [PubMed]
- Hastie, K.M.; Zandonatti, M.A.; Kleinfelter, L.M.; Heinrich, M.L.; Rowland, M.M.; Chandran, K.; Branco, L.M.; Robinson, J.E.; Garry, R.F.; Saphire, E.O. Structural basis for antibody-mediated neutralization of lassa virus. Science 2017, 356, 923–928. [Google Scholar] [CrossRef] [PubMed]
- Gallaher, W.R. Similar structural models of the transmembrane proteins of ebola and avian sarcoma viruses. Cell 1996, 85, 477–478. [Google Scholar] [CrossRef]
- Gallaher, W.R.; Ball, J.M.; Garry, R.F.; Griffin, M.C.; Montelaro, R.C. A general model for the transmembrane proteins of hiv and other retroviruses. AIDS Res. Hum. Retrovir. 1989, 5, 431–440. [Google Scholar] [CrossRef]
- Gallaher, W.R.; DiSimone, C.; Buchmeier, M.J. The viral transmembrane superfamily: Possible divergence of arenavirus and filovirus glycoproteins from a common rna virus ancestor. BMC Microbiol. 2001, 1, 1. [Google Scholar] [CrossRef]
- Garry, C.E.; Garry, R.F. Proteomics computational analyses suggest that the carboxyl terminal glycoproteins of bunyaviruses are class ii viral fusion protein (beta-penetrenes). Theor. Biol. Med Model. 2004, 1, 10. [Google Scholar] [CrossRef]
- Garry, C.E.; Garry, R.F. Proteomics computational analyses suggest that baculovirus gp64 superfamily proteins are class iii penetrenes. Virol. J. 2008, 5, 28. [Google Scholar] [CrossRef]
- Garry, C.E.; Garry, R.F. Proteomics computational analyses suggest that the bornavirus glycoprotein is a class iii viral fusion protein (gamma penetrene). Virol. J. 2009, 6, 145. [Google Scholar] [CrossRef] [PubMed]
- Pearson, W. Lalign. Available online: https://embnet.vital-it.ch/software/LALIGN_form.html (accessed on 7 July 2019).
- Huang, X.; Miller, W. A time-efficient, linear-space local similarity algorithm. Adv. Appl. Math. 1991, 12, 337–357. [Google Scholar] [CrossRef] [Green Version]
- Madeira, F.; Park, Y.M.; Lee, J.; Buso, N.; Gur, T.; Madhusoodanan, N.; Basutkar, P.; Tivey, A.R.N.; Potter, S.C.; Finn, R.D.; et al. The embl-ebi search and sequence analysis tools apis in 2019. Nucleic Acids Res. 2019, 47, w636–w641. [Google Scholar] [CrossRef] [PubMed]
- Yachdav, G.; Kloppmann, E.; Kajan, L.; Hecht, M.; Goldberg, T.; Hamp, T.; Honigschmid, P.; Schafferhans, A.; Roos, M.; Bernhofer, M.; et al. Predictprotein--an open resource for online prediction of protein structural and functional features. Nucleic Acids Res. 2014, 42, w337–w343. [Google Scholar] [CrossRef]
- Hofmann, K.; Stoffel, W. Tmpred. Available online: https://embnet.vital-it.ch/software/TMPRED_form.html (accessed on 7 July 2019).
- Hofmann, K.; Stoffel, W. Tmbase—A database of membrane spanning proteins segments. Biol. Chem. Hoppe-Seyler 1993, 374, 166. [Google Scholar]
- White, S.H.; Snider, C.; Jaysinghe, S.; Kim, J. Membrane Protein Explorer Version 2.2a. Available online: http://blanco.biomol.uci.edu/mpex/ 2003 (accessed on 7 July 2019).
- Almagro Armenteros, J.J.; Tsirigos, K.D.; Sonderby, C.K.; Petersen, T.N.; Winther, O.; Brunak, S.; von Heijne, G.; Nielsen, H. Signalp 5.0 improves signal peptide predictions using deep neural networks. Nat. Biotechnol. 2019, 37, 420–423. [Google Scholar] [CrossRef] [PubMed]
- Steentoft, C.; Vakhrushev, S.Y.; Joshi, H.J.; Kong, Y.; Vester-Christensen, M.B.; Schjoldager, K.T.; Lavrsen, K.; Dabelsteen, S.; Pedersen, N.B.; Marcos-Silva, L.; et al. Precision mapping of the human o-galnac glycoproteome through simplecell technology. EMBO J. 2013, 32, 1478–1488. [Google Scholar] [CrossRef] [PubMed]
- Briknarova, K.; Thomas, C.J.; York, J.; Nunberg, J.H. Structure of a zinc-binding domain in the junin virus envelope glycoprotein. J. Biol Chem 2011, 286, 1528–1536. [Google Scholar] [CrossRef] [PubMed]
- York, J.; Nunberg, J.H. A novel zinc-binding domain is essential for formation of the functional junin virus envelope glycoprotein complex. J. Virol. 2007, 81, 13385–13391. [Google Scholar] [CrossRef] [PubMed]
- Saez-Cirion, A.; Gomara, M.J.; Agirre, A.; Nieva, J.L. Pre-transmembrane sequence of ebola glycoprotein. Interfacial hydrophobicity distribution and interaction with membranes. FEBS Lett. 2003, 533, 47–53. [Google Scholar] [CrossRef]
- Suarez, T.; Gallaher, W.R.; Agirre, A.; Goni, F.M.; Nieva, J.L. Membrane interface-interacting sequences within the ectodomain of the human immunodeficiency virus type 1 envelope glycoprotein: Putative role during viral fusion. J. Virol. 2000, 74, 8038–8047. [Google Scholar] [CrossRef] [PubMed]
- Sainz, B., Jr.; Rausch, J.M.; Gallaher, W.R.; Garry, R.F.; Wimley, W.C. The aromatic domain of the coronavirus class i viral fusion protein induces membrane permeabilization: Putative role during viral entry. Biochemistry 2005, 44, 947–958. [Google Scholar] [CrossRef] [PubMed]
- Abraham, J.; Corbett, K.D.; Farzan, M.; Choe, H.; Harrison, S.C. Structural basis for receptor recognition by new world hemorrhagic fever arenaviruses. Nat. Struct. Mol. Biol. 2010, 17, 438–444. [Google Scholar] [CrossRef] [PubMed]
- Hastie, K.M.; Igonet, S.; Sullivan, B.M.; Legrand, P.; Zandonatti, M.A.; Robinson, J.E.; Garry, R.F.; Rey, F.A.; Oldsone, M.B.; Saphire, E.O. Crystal structure of the prefusion surface glycoprotein of the prototypic arenavirus lcmv. Nat. Struct. Mol. Biol. 2016, 6, 513–521. [Google Scholar] [CrossRef] [PubMed]
- Cao, W.; Henry, M.D.; Borrow, P.; Yamada, H.; Elder, J.H.; Ravkov, E.V.; Nichol, S.T.; Compans, R.W.; Campbell, K.P.; Oldstone, M.B. Identification of alpha-dystroglycan as a receptor for lymphocytic choriomeningitis virus and lassa fever virus. Science 1998, 282, 2079–2081. [Google Scholar] [CrossRef] [PubMed]
- Spiropoulou, C.F.; Kunz, S.; Rollin, P.E.; Campbell, K.P.; Oldstone, M.B. New world arenavirus clade c, but not clade a and b viruses, utilizes alpha-dystroglycan as its major receptor. J. Virol. 2002, 76, 5140–5146. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.E.; Fusco, M.L.; Hessell, A.J.; Oswald, W.B.; Burton, D.R.; Saphire, E.O. Structure of the ebola virus glycoprotein bound to an antibody from a human survivor. Nature 2008, 454, 177–182. [Google Scholar] [CrossRef] [PubMed]
- Hastie, K.M.; Kimberlin, C.R.; Zandonatti, M.A.; MacRae, I.J.; Saphire, E.O. Structure of the lassa virus nucleoprotein reveals a dsrna-specific 3′ to 5′ exonuclease activity essential for immune suppression. Proc. Natl. Acad. Sci. USA 2011, 108, 2396–2401. [Google Scholar] [CrossRef] [PubMed]
- Hastie, K.M.; Liu, T.; Li, S.; King, L.B.; Ngo, N.; Zandonatti, M.A.; Woods, V.L., Jr.; de la Torre, J.C.; Saphire, E.O. Crystal structure of the lassa virus nucleoprotein-rna complex reveals a gating mechanism for rna binding. Proc. Natl. Acad. Sci. USA 2011, 108, 19365–19370. [Google Scholar] [CrossRef]
- Wilson, I.A.; Skehel, J.J.; Wiley, D.C. Structure of the haemagglutinin membrane glycoprotein of influenza virus at 3 a resolution. Nature 1981, 289, 366–373. [Google Scholar] [CrossRef]
- Chan, D.C.; Fass, D.; Berger, J.M.; Kim, P.S. Core structure of gp41 from the hiv envelope glycoprotein. Cell 1997, 89, 263–273. [Google Scholar] [CrossRef]
- Weissenhorn, W.; Dessen, A.; Harrison, S.C.; Skehel, J.J.; Wiley, D.C. Atomic structure of the ectodomain from hiv-1 gp41. Nature 1997, 387, 426–430. [Google Scholar] [CrossRef] [PubMed]
- Kowalski, M.; Potz, J.; Basiripour, L.; Dorfman, T.; Haseltine, W.; Sodroski, J. Attenuation of hiv-1 cytopathic effect by mutation affecting the transmembrane glycoprotein. J. Virol. 1991, 65, 281–291. [Google Scholar] [PubMed]
- Gallaher, W.R.; Garry, R.F. Model of the Pre-Insertion Region of the Spike (s2) Fusion Glycoprotein of the Human Sars Coronavirus: Implications for Antiviral Therapeutics. Available online: http://www.virology.net/sars/s2model.html (accessed on 7 July 2019).
- Liu, S.; Xiao, G.; Chen, Y.; He, Y.; Niu, J.; Escalante, C.R.; Xiong, H.; Farmar, J.; Debnath, A.K.; Tien, P.; et al. Interaction between heptad repeat 1 and 2 regions in spike protein of sars-associated coronavirus: Implications for virus fusogenic mechanism and identification of fusion inhibitors. Lancet 2004, 363, 938–947. [Google Scholar] [CrossRef]
- Tripet, B.; Howard, M.W.; Jobling, M.; Holmes, R.K.; Holmes, K.V.; Hodges, R.S. Structural characterization of the sars-coronavirus spike s fusion protein core. J. Biol. Chem. 2004, 279, 20836–20849. [Google Scholar] [CrossRef] [PubMed]
- Baker, K.A.; Dutch, R.E.; Lamb, R.A.; Jardetzky, T.S. Structural basis for paramyxovirus-mediated membrane fusion. Mol. Cell 1999, 3, 309–319. [Google Scholar] [CrossRef]
- Rey, F.A.; Heinz, F.X.; Mandl, C.; Kunz, C.; Harrison, S.C. The envelope glycoprotein from tick-borne encephalitis virus at 2 a resolution. Nature 1995, 375, 291–298. [Google Scholar] [CrossRef]
- Lescar, J.; Roussel, A.; Wien, M.W.; Navaza, J.; Fuller, S.D.; Wengler, G.; Wengler, G.; Rey, F.A. The fusion glycoprotein shell of semliki forest virus: An icosahedral assembly primed for fusogenic activation at endosomal ph. Cell 2001, 105, 137–148. [Google Scholar] [CrossRef]
- Heldwein, E.E.; Lou, H.; Bender, F.C.; Cohen, G.H.; Eisenberg, R.J.; Harrison, S.C. Crystal structure of glycoprotein b from herpes simplex virus 1. Science 2006, 313, 217–220. [Google Scholar] [CrossRef]
- Roche, S.; Bressanelli, S.; Rey, F.A.; Gaudin, Y. Crystal structure of the low-ph form of the vesicular stomatitis virus glycoprotein g. Science 2006, 313, 187–191. [Google Scholar] [CrossRef]
- Roche, S.; Rey, F.A.; Gaudin, Y.; Bressanelli, S. Structure of the prefusion form of the vesicular stomatitis virus glycoprotein g. Science 2007, 315, 843–848. [Google Scholar] [CrossRef]
- Misseri, Y.; Labesse, G.; Bucheton, A.; Terzian, C. Comparative sequence analysis and predictions for the envelope glycoproteins of insect endogenous retroviruses. Trends Microbiol. 2003, 11, 253–256. [Google Scholar] [CrossRef]
- Stenglein, M.D.; Jacobson, E.R.; Chang, L.W.; Sanders, C.; Hawkins, M.G.; Guzman, D.S.; Drazenovich, T.; Dunker, F.; Kamaka, E.K.; Fisher, D.; et al. Widespread recombination, reassortment, and transmission of unbalanced compound viral genotypes in natural arenavirus infections. PLoS Pathog. 2015, 11, e1004900. [Google Scholar] [CrossRef]
- Hugot, J.P.; Gonzalez, J.P.; Denys, C. Evolution of the old world arenaviridae and their rodent hosts: Generalized host-transfer or association by descent? Infect. Genet. Evol. 2001, 1, 13–20. [Google Scholar] [CrossRef]
- Forni, D.; Pontremoli, C.; Pozzoli, U.; Clerici, M.; Cagliani, R.; Sironi, M. Ancient evolution of mammarenaviruses: Adaptation via changes in the l protein and no evidence for host-virus codivergence. Genome Biol. Evol. 2018, 10, 863–874. [Google Scholar] [CrossRef]
- Ramsden, C.; Holmes, E.C.; Charleston, M.A. Hantavirus evolution in relation to its rodent and insectivore hosts: No evidence for codivergence. Mol. Biol. Evol. 2009, 26, 143–153. [Google Scholar] [CrossRef]
- Coulibaly-N’Golo, D.; Allali, B.; Kouassi, S.K.; Fichet-Calvet, E.; Becker-Ziaja, B.; Rieger, T.; Olschlager, S.; Dosso, H.; Denys, C.; Ter Meulen, J.; et al. Novel arenavirus sequences in Hylomyscus sp. and Mus (Nannomys) setulosus from cote d’ivoire: Implications for evolution of arenaviruses in africa. PLoS ONE 2011, 6, e20893. [Google Scholar] [CrossRef]






© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Garry, C.E.; Garry, R.F. Proteomics Computational Analyses Suggest that the Antennavirus Glycoprotein Complex Includes a Class I Viral Fusion Protein (α-Penetrene) with an Internal Zinc-Binding Domain and a Stable Signal Peptide. Viruses 2019, 11, 750. https://doi.org/10.3390/v11080750
Garry CE, Garry RF. Proteomics Computational Analyses Suggest that the Antennavirus Glycoprotein Complex Includes a Class I Viral Fusion Protein (α-Penetrene) with an Internal Zinc-Binding Domain and a Stable Signal Peptide. Viruses. 2019; 11(8):750. https://doi.org/10.3390/v11080750
Chicago/Turabian StyleGarry, Courtney E., and Robert F. Garry. 2019. "Proteomics Computational Analyses Suggest that the Antennavirus Glycoprotein Complex Includes a Class I Viral Fusion Protein (α-Penetrene) with an Internal Zinc-Binding Domain and a Stable Signal Peptide" Viruses 11, no. 8: 750. https://doi.org/10.3390/v11080750
APA StyleGarry, C. E., & Garry, R. F. (2019). Proteomics Computational Analyses Suggest that the Antennavirus Glycoprotein Complex Includes a Class I Viral Fusion Protein (α-Penetrene) with an Internal Zinc-Binding Domain and a Stable Signal Peptide. Viruses, 11(8), 750. https://doi.org/10.3390/v11080750