A Cut above the Rest: Characterization of the Assembly of a Large Viral Icosahedral Capsid
Abstract
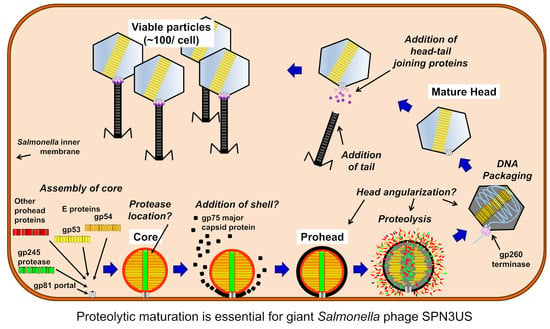
:1. Introduction
2. Materials and Methods
2.1. Isolation and Genome Sequencing of SPN3US Protease Mutant Phages
2.2. Purification and TEM of SPN3US Amber Mutant Particles
2.3. Mass Spectrometry Analyses of SPN3US Precursor Particles
2.4. Cloning, Expression, and Purification of Truncated Forms of SPN3US gp245
2.5. Transmission Electron Microscopy (TEM) of SPN3US-Infected Salmonella
2.6. Growth of SPN3US in Liquid Culture
3. Results and Discussion
3.1. Genome Sequencing of Mutant Phages Confirms the Essential Status of the SPN3US Protease
3.2. SPN3US Protease Mutant 245(am59) Undergoes Normal Head Maturation under Amber Suppressing Conditions
3.3. Immature SPN3US Proheads Accumulate in Protease Mutants Grown under Non-Amber Suppressing Conditions
3.4. SPN3US Proheads in Protease Mutant Infections Do Not Undergo Normal Proteolytic Maturation
3.5. The C-Terminus of the SPN3US Protease Likely Has a Role in Targeting the Protease into the Prohead
3.6. SPN3US Proheads Assemble on the Inner Salmonella Membrane
3.7. Maturation Results in Major Morphological Changes in WT SPN3US Proheads
3.8. SPN3US Virion Formation and Infection Progress Rapidly
3.9. SPN3US Infection Alters Salmonella Cell Morphology
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Fokine, A.; Rossmann, M.G. Molecular architecture of tailed double-stranded DNA phages. Bacteriophage 2014, 4, e28281. [Google Scholar] [CrossRef] [Green Version]
- Earnshaw, W.C.; Casjens, S.R. DNA packaging by the double-stranded DNA bacteriophages. Cell 1980, 21, 319–331. [Google Scholar] [CrossRef]
- Hendrix, R.W.; Johnson, J.E. Bacteriophage HK97 capsid assembly and maturation. In Viral Molecular Machines; Rossmann, M.G., Rao, V.B., Eds.; Springer: Boston, MA, USA, 2012; pp. 351–363. [Google Scholar] [CrossRef]
- Rao, V.; Black, L. Structure and assembly of bacteriophage T4 head. Virol. J. 2010, 7, 356. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aksyuk, A.A.; Rossmann, M.G. Bacteriophage assembly. Viruses 2011, 3, 172–203. [Google Scholar] [CrossRef] [PubMed]
- Prevelige, P.E.; Cortines, J.R. Phage assembly and the special role of the portal protein. Curr. Opin. Virol. 2018, 31, 66–73. [Google Scholar] [CrossRef]
- Fokine, A.; Leiman, P.G.; Shneider, M.M.; Ahvazi, B.; Boeshans, K.M.; Steven, A.C.; Black, L.W.; Mesyanzhinov, V.V.; Rossmann, M.G. Structural and functional similarities between the capsid proteins of bacteriophages T4 and HK97 point to a common ancestry. Proc. Natl. Acad. Sci. USA 2005, 102, 7163–7168. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pietilä, M.K.; Laurinmäki, P.; Russell, D.A.; Ko, C.-C.; Jacobs-Sera, D.; Hendrix, R.W.; Bamford, D.H.; Butcher, S.J. Structure of the archaeal head-tailed virus HSTV-1 completes the HK97 fold story. Proc. Natl. Acad. Sci. USA 2013, 110, 10604. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Selvarajan Sigamani, S.; Zhao, H.; Kamau, Y.N.; Baines, J.D.; Tang, L. The structure of the herpes simplex virus DNA-packaging terminase pUL15 nuclease domain suggests an evolutionary lineage among eukaryotic and prokaryotic viruses. J. Virol. 2013, 87, 7140–7148. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.; Mushegian, A. Displacements of prohead protease genes in the late operons of double-stranded-DNA bacteriophages. J. Bacteriol. 2004, 186, 4369–4375. [Google Scholar] [CrossRef] [Green Version]
- Cheng, H.; Shen, N.; Pei, J.; Grishin, N.V. Double-stranded DNA bacteriophage prohead protease is homologous to herpesvirus protease. Protein Sci. 2004, 13, 2260–2269. [Google Scholar] [CrossRef] [Green Version]
- Fokine, A.; Rossmann, M.G. Common evolutionary origin of procapsid proteases, phage tail tubes, and tubes of bacterial Type VI secretion systems. Structure 2016, 24, 1928–1935. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Duda, R.L.; Oh, B.; Hendrix, R.W. Functional domains of the HK97 capsid maturation protease and the mechanisms of protein encapsidation. J. Mol. Biol. 2013, 425, 2765–2781. [Google Scholar] [CrossRef] [Green Version]
- Showe, M.K.; Isobe, E.; Onorato, L. Bacteriophage T4 prehead proteinase: I. Purification and properties of a bacteriophage enzyme which cleaves the capsid precursor proteins. J. Mol. Biol. 1976, 107, 35–54. [Google Scholar] [CrossRef]
- Showe, M.K.; Isobe, E.; Onorato, L. Bacteriophage T4 prehead proteinase: II. Its cleavage from the product of gene 21 and regulation in phage-infected cells. J. Mol. Biol. 1976, 107, 55–69. [Google Scholar] [CrossRef]
- Medina, E.; Wieczorek, D.; Medina, E.M.; Yang, Q.; Feiss, M.; Catalano, C.E. Assembly and maturation of the bacteriophage lambda procapsid: GpC is the viral protease. J. Mol. Biol. 2010, 401, 813–830. [Google Scholar] [CrossRef]
- Onorato, L.; Showe, M.K. Gene 21 protein-dependent proteolysis in vitro of purified gene 22 product of bacteriophage T4. J. Mol. Biol. 1975, 92, 395–412. [Google Scholar] [CrossRef]
- Keller, B.; Maeder, M.; Becker-Laburte, C.; Kellenberger, E.; Bickle, T.A. Amber mutants in gene 67 of phage T4. Effects on formation and shape determination of the head. J. Mol. Biol. 1986, 190, 83–95. [Google Scholar] [CrossRef]
- Keller, B.; Dubochet, J.; Adrian, M.; Maeder, M.; Wurtz, M.; Kellenberger, E. Length and shape variants of the bacteriophage T4 head: Mutations in the scaffolding core genes 68 and 22. J. Virol. 1988, 62, 2960–2969. [Google Scholar] [CrossRef] [Green Version]
- Hinton, D.M. Transcriptional control in the prereplicative phase of T4 development. Virol. J. 2010, 7, 289. [Google Scholar] [CrossRef] [Green Version]
- Goff, C.G. Bacteriophage T4 alt gene maps between genes 30 and 54. J. Virol. 1979, 29, 1232–1234. [Google Scholar] [CrossRef] [Green Version]
- Black, L.W.; Brown, D.T. Head morphologies in bacteriophage T4 head and internal protein mutant infections. J. Virol. 1976, 17, 894–905. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Caldentey, J.; Lepault, J.; Kellenberger, E. Isolation and reassembly of bacteriophage T4 core proteins. J. Mol. Biol. 1987, 195, 637–647. [Google Scholar] [CrossRef]
- Mullaney, J.M.; Black, L.W. Capsid targeting sequence targets foreign proteins into bacteriophage T4 and permits proteolytic processing. J. Mol. Biol. 1996, 261, 372–385. [Google Scholar] [CrossRef] [PubMed]
- Hendrix, R.W. Jumbo bacteriophages. In Lesser Known Large dsDNA Viruses; Van Etten, J., Ed.; Springer: Berlin/Heidelberg, Germany, 2009; Volume 328, pp. 229–240. [Google Scholar]
- Mesyanzhinov, V.V.; Robben, J.; Grymonprez, B.; Kostyuchenko, V.A.; Bourkaltseva, M.V.; Sykilinda, N.N.; Krylov, V.N.; Volckaert, G. The genome of bacteriophage PhiKZ of Pseudomonas aeruginosa. J. Mol. Biol. 2002, 317, 1–19. [Google Scholar] [CrossRef]
- Lee, J.-H.; Shin, H.; Kim, H.; Ryu, S. Complete genome sequence of Salmonella bacteriophage SPN3US. J. Virol. 2011, 85, 13470–13471. [Google Scholar] [CrossRef] [Green Version]
- Thomas, J.A.; Benítez Quintana, A.D.; Bosch, M.A.; Coll De Peña, A.; Aguilera, E.; Coulibaly, A.; Wu, W.; Osier, M.V.; Hudson, A.O.; Weintraub, S.T.; et al. Identification of essential genes in the Salmonella phage SPN3US reveals novel insights into giant phage head structure and assembly. J. Virol. 2016, 90, 10284–10298. [Google Scholar] [CrossRef] [Green Version]
- Ali, B.; Desmond, M.I.; Mallory, S.A.; Benitez, A.D.; Buckley, L.J.; Weintraub, S.T.; Osier, M.V.; Black, L.W.; Thomas, J.A. To Be or Not To Be T4: Evidence of a Complex Evolutionary Pathway of Head Structure and Assembly in Giant Salmonella Virus SPN3US. Front. Microbiol. 2017, 8, 2251. [Google Scholar] [CrossRef] [Green Version]
- Thomas, J.A.; Weintraub, S.T.; Hakala, K.; Serwer, P.; Hardies, S.C. Proteome of the large Pseudomonas myovirus 201φ2-1: Delineation of proteolytically processed virion proteins. Mol. Cell. Proteom. 2010, 9, 940–951. [Google Scholar] [CrossRef] [Green Version]
- Thomas, J.A.; Weintraub, S.T.; Wu, W.; Winkler, D.C.; Cheng, N.; Steven, A.C.; Black, L.W. Extensive proteolysis of head and inner body proteins by a morphogenetic protease in the giant Pseudomonas aeruginosa phage φKZ. Mol. Microbiol. 2012, 84, 324–339. [Google Scholar] [CrossRef] [Green Version]
- Qiu, D.; Damron, F.H.; Mima, T.; Schweizer, H.P.; Yu, H.D. PBAD-based shuttle vectors for functional analysis of toxic and highly regulated genes in Pseudomonas and Burkholderia spp. and other bacteria. Appl. Environ. Microbiol. 2008, 74, 7422–7426. [Google Scholar] [CrossRef] [Green Version]
- Thomas, J.A.; Black, L.W. Mutational analysis of the Pseudomonas aeruginosa myovirus φKZ morphogenetic protease gp175. J. Virol. 2013, 87, 8713–8725. [Google Scholar] [CrossRef] [Green Version]
- Ackermann, H.-W.; Tiekotter, K.L. Murphy’s law-if anything can go wrong, it will: Problems in phage electron microscopy. Bacteriophage 2012, 2, 122–129. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- van Driel, R.; Traub, F.; Showe, M.K. Probable localization of the bacteriophage T4 prehead proteinase zymogen in the center of the prehead core. J. Virol. 1980, 36, 220–223. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weintraub, S.T.; Redzuan, N.H.M.; Barton, M.K.; Md Amin, N.A.; Desmond, M.I.; Adams, L.E.; Ali, B.; Pardo, S.; Molleur, D.; Wu, W.; et al. Global proteomic profiling of Salmonella infection by a giant phage. J. Virol. 2019, 93. [Google Scholar] [CrossRef] [Green Version]
- Mullaney, J.M.; Black, L.W. Bacteriophage T4 capsid packaging and unpackaging of DNA and proteins. Methods Mol. Biol. 2014, 1108, 69–85. [Google Scholar] [CrossRef]
- Miller, E.S.; Kutter, E.; Mosig, G.; Arisaka, F.; Kunisawa, T.; Ruger, W. Bacteriophage T4 Genome. Microbiol. Mol. Biol. Rev. 2003, 67, 86–156. [Google Scholar] [CrossRef] [Green Version]
- Black, L.W.; Showe, M.K.; Steven, A.C. Morphogenesis of the T4 head. In Molecular Biology of Bacteriophage T4; Karam, J.D., Ed.; ASM Press: Washington, DC, USA, 1994; pp. 218–258. [Google Scholar]
- Sokolova, O.S.; Shaburova, O.V.; Pechnikova, E.V.; Shaytan, A.K.; Krylov, S.V.; Kiselev, N.A.; Krylov, V.N. Genome packaging in EL and Lin68, two giant PhiKZ-like bacteriophages of P. aeruginosa. Virology 2014, 468–470, 472–478. [Google Scholar] [CrossRef] [Green Version]
- Krylov, V.N.; Smirnova, T.A.; Minenkova, I.B.; Plotnilova, T.G.; Zhazikov, I.Z.; Khrenova, E.A. Pseudomonas bacteriophage PhiKZ contains an inner body in its capsid. Can. J. Microbiol. 1984, 30, 758–762. [Google Scholar] [CrossRef]
- Thomas, J.A.; Rolando, M.R.; Carroll, C.A.; Shen, P.S.; Belnap, D.M.; Weintraub, S.T.; Serwer, P.; Hardies, S.C. Characterization of Pseudomonas chlororaphis myovirus 201φ2-1 via genomic sequencing, mass spectrometry, and electron microscopy. Virology 2008, 376, 330–338. [Google Scholar] [CrossRef] [Green Version]
- Wu, W.; Thomas, J.A.; Cheng, N.; Black, L.W.; Steven, A.C. Bubblegrams reveal the inner body structure of φKZ. Science 2012, 335, 182. [Google Scholar] [CrossRef] [Green Version]
- Lavysh, D.; Sokolova, M.; Slashcheva, M.; Förstner, K.U.; Severinov, K. Transcription profiling of Bacillus subtilis cells infected with AR9, a giant phage encoding two multisubunit RNA polymerases. mBio 2017, 8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Robinow, C.; Kellenberger, E. The bacterial nucleoid revisited. Microbiol. Rev. 1994, 58, 211–232. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eltsov, M.; Zuber, B. Transmission electron microscopy of the bacterial nucleoid. J. Struct. Biol. 2006, 156, 246–254. [Google Scholar] [CrossRef] [PubMed]








| Mutant | Reference Position 1 | Gene Name | DNA Change 2,3,4 | Amino Acid Change 5 | Impact | Function/Comment |
|---|---|---|---|---|---|---|
| am59 | 29457 | SPN3US_0034 | c.444G>A | p.Q148Q | Synonymous | Non-virion RNAP β |
| 46070 | SPN3US_0047 | c.1172G>A | p.G391D | Non Synonymous | Low abundance head protein | |
| 59923 | SPN3US_0061 | c.211G>A | p.V71M | Non Synonymous | Putative neck protein | |
| 85238 | SPN3US_0086 | c.261A>G | p.E87E | Synonymous | Non-virion protein | |
| 114280 | SPN3US_0135 | c.222C>T | p.D74D | Synonymous | Non-virion protein | |
| 122374 | SPN3US_0145 | c.857C>T | p.P286L | Non Synonymous | Low abundance head protein | |
| 147028 | SPN3US_0168 | c.3692T>C | p.L1231P | Non Synonymous | Tail fiber protein | |
| 186363 | SPN3US_0213 | c.187C>T | p.R63C | Non Synonymous | Non-virion protein | |
| 210826 | SPN3US_0239 | c.1502C>T | p.A501V | Non Synonymous | Tape measure protein | |
| 221150 | SPN3US_0245 | c.343C>T | p.Q115. | Nonsense | Prohead protease | |
| 230553 | SPN3US_0258 | c.395C>T | p.T132M | Non Synonymous | Tail or neck protein | |
| 239933 * | g.239933insT | Mutation in non-coding region | ||||
| am66 | 77962 * | g.77962insT | Mutation in non-coding region | |||
| 87715 * | g.87715insC | Mutation in non-coding region | ||||
| 93451 * | g.93451insT | Mutation in non-coding region | ||||
| 135828 | SPN3US_0156 | c.404G>A | p.G135E | Non-synonymous | Non-virion protein | |
| 180411 | SPN3US_0204 | c.395C>T | p.A132V | Non-synonymous | Non-virion protein | |
| 200944 * | g.200944insT | Mutation in non-coding region | ||||
| 221528 | SPN3US_0245 | c.721C>T | p.Q241. | Nonsense | Prohead protease | |
| 223871 | SPN3US_0250 | c.70C>A | p.R24S | Non-synonymous | Non-virion protein | |
| 239933 * | g.239933insT | Mutation in non-coding region |
| SPN3US | Salmonella | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| SPN3US Phage/Mutant | Experiment Abbreviation | Purification 1 | Refractive Index | Buoyant Density, g/mL | No. of Proteins Identified | Total PSM 2 | No. of Proteins Identified | Total PSM 2 | Salmonella/SPN3US PSM 2, % |
| Wild-type 3 | WT | CsCl step gradient 4 | 1.3735 (Lower, “junk” band = 1.3695/1.3694) | 1.42 (1.38) | 86 | 8513 | 28 | 470 | 5.5 |
| 245(am59) | am59-supD | CsCl step gradient 4 | 1.3725 | 1.41 | 62 | 2638 | 42 | 410 | 15.54 |
| am59-supF (tailed) | CsCl step and buoyant density gradients 5 | 1.3735 | 1.42 | 88 | 9689 | 20 | 72 | 0.74 | |
| am59-supF (heads) | CsCl step and buoyant density gradients 5 | 1.3749 | 1.43 | 74 | 9214 | 16 | 80 | 0.87 | |
| am59-sup0-D | Differential concentration only | NA | NA | 90 | 4018 | 188 | 2232 | 55.55 | |
| am59-sup0-SB | CsCl step bottom band 4 | 1.3685 | 1.37 | 66 | 186 | ||||
| am59-sup0-ST | CsCl step top band 4 | 1.3636 | 1.32 | ND | ND | ND | ND | ND | |
| am59-sup0-a | CsCl step and buoyant density gradients 5 | 1.3650 | 1.33 | 75 | 4405 | 36 | 361 | 8.20 | |
| am59-sup0-b | CsCl step and buoyant density gradients 5 | 1.3640 | 1.32 | 101 | 11326 | 80 | 857 | 7.57 | |
| am59-sup0-glut | CsCl step and buoyant density gradients 6 | 1.3640 | 1.32 | ND | ND | ND | ND | ND | |
| 245(am66) | am66-sup0 | CsCl step and buoyant density gradients 5 | 1.3638 | 1.32 | 113 | 11102 | 83 | 640 | 5.76 |
| Amber Suppressing Growth Condition (PSM/M) 1 | Non-Amber suppressing Growth Conditions (PSM/M) 1 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| gp | Function | Mass, kDa (Proc. 2) | WT | am59-supD | am59-supF (Tailed) | am59-supF (Heads) | am59-sup0-D | am59-sup0-a | am59-sup0-b | am66-sup0 |
| Highly abundant proteins | ||||||||||
| 75 | Major capsid | 83.9 (70.4) | 22.61 | 9.26 | 22.78 | 33.51 | 10.60 | 15.11 | 33.06 | 31.33 |
| 22 | Scaffold candidate | 67.1 | 0.00 | 0.40 | 0.00 | 0.00 | 8.91 | 16.99 | 21.33 | 26.87 |
| 256 | Tail sheath | 75.7 | 8.84 | 3.36 | 8.27 | 0.94 | 2.60 | 3.71 | 9.82 | 11.72 |
| 53 | E protein | 45.2 (31.5) | 21.02 | 5.65 | 24.86 | 29.68 | 9.98 | 5.55 | 14.12 | 10.53 |
| 54 | E protein | 45.1 (31.9) | 20.16 | 3.64 | 21.00 | 29.31 | 7.27 | 5.01 | 10.22 | 9.02 |
| Low abundance proteins | ||||||||||
| 81 | Portal | 100.2 (72.3) | 1.29 | 0.39 | 1.22 | 2.09 | 0.06 | 0.11 | 0.76 | 0.82 |
| 245 | Prohead protease | 30.7 (23.4) | 1.07 | 0.13 | 1.11 | 1.11 | 0.00 | 0.00 | 0.07 | 0.07 |
| 61 | Possible neck | 58.3 | 0.60 | 0.10 | 0.50 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 |
| 62 | Possible neck | 52.0 | 0.85 | 0.19 | 0.77 | 0.04 | 0.00 | 0.00 | 0.00 | 0.00 |
| 64 | Possible neck | 48.9 | 0.41 | 0.00 | 0.41 | 0.00 | 0.00 | 0.04 | 0.37 | 0.20 |
| Protein | Cleavage Site | Residues | Peptide Sequence 1 | No. of Peptide Spectrum Matches | |
|---|---|---|---|---|---|
| am59-sup0-b | am66-sup0 | ||||
| gp75, MCP | ATE-130 | 124–150 | TPLVATEYYTNKDLDKNLGLTWTLNVR | 53 | 60 |
| 124–139 | TPLVATEYYTNKDLDK | 14 | 14 | ||
| 124–135 | TPLVATEYYTNK | 12 | 10 | ||
| 94–135 | VAAESAKPEYASYMPIAISGAASLAADYGKTPLVATEYYTNK | 4 | 9 | ||
| 94–150 | VAAESAKPEYASYMPIAISGAASLAADYGKTPLVATEYYTNKDLDKNLGLTWTLNVR | 0 | 5 | ||
| 94–139 | VAAESAKPEYASYMPIAISGAASLAADYGKTPLVATEYYTNKDLDK | 0 | 1 | ||
| 124–148 | TPLVATEYYTNKDLDKNLGLTWTLN | 1 | 1 | ||
| 124–145 | TPLVATEYYTNKDLDKNLGLTW | 1 | 1 | ||
| 124–138 | TPLVATEYYTNKDLD | 1 | 1 | ||
| 124–136 | TPLVATEYYTNKD | 1 | 1 | ||
| gp81, Portal | ATE-161 (expected maturation Site: AQE-254) | 136–164 | GASPVLLISDTGFDELFGLKPSVATESLR | 1 | 2 |
| 136–165 | GASPVLLISDTGFDELFGLKPSVATESLRR | 1 | 2 | ||
| gp53, E protein | AQE-125 | 122–137 | AAQEGWKETLKDLFER | 10 | 5 |
| 122–132 | AAQEGWKETLK | 2 | 1 | ||
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Reilly, E.R.; Abajorga, M.K.; Kiser, C.; Mohd Redzuan, N.H.; Haidar, Z.; Adams, L.E.; Diaz, R.; Pinzon, J.A.; Hudson, A.O.; Black, L.W.; et al. A Cut above the Rest: Characterization of the Assembly of a Large Viral Icosahedral Capsid. Viruses 2020, 12, 725. https://doi.org/10.3390/v12070725
Reilly ER, Abajorga MK, Kiser C, Mohd Redzuan NH, Haidar Z, Adams LE, Diaz R, Pinzon JA, Hudson AO, Black LW, et al. A Cut above the Rest: Characterization of the Assembly of a Large Viral Icosahedral Capsid. Viruses. 2020; 12(7):725. https://doi.org/10.3390/v12070725
Chicago/Turabian StyleReilly, Erin R., Milky K. Abajorga, Cory Kiser, Nurul Humaira Mohd Redzuan, Zein Haidar, Lily E. Adams, Randy Diaz, Juliana A. Pinzon, André O. Hudson, Lindsay W. Black, and et al. 2020. "A Cut above the Rest: Characterization of the Assembly of a Large Viral Icosahedral Capsid" Viruses 12, no. 7: 725. https://doi.org/10.3390/v12070725
APA StyleReilly, E. R., Abajorga, M. K., Kiser, C., Mohd Redzuan, N. H., Haidar, Z., Adams, L. E., Diaz, R., Pinzon, J. A., Hudson, A. O., Black, L. W., Hsia, R.-C., Weintraub, S. T., & Thomas, J. A. (2020). A Cut above the Rest: Characterization of the Assembly of a Large Viral Icosahedral Capsid. Viruses, 12(7), 725. https://doi.org/10.3390/v12070725