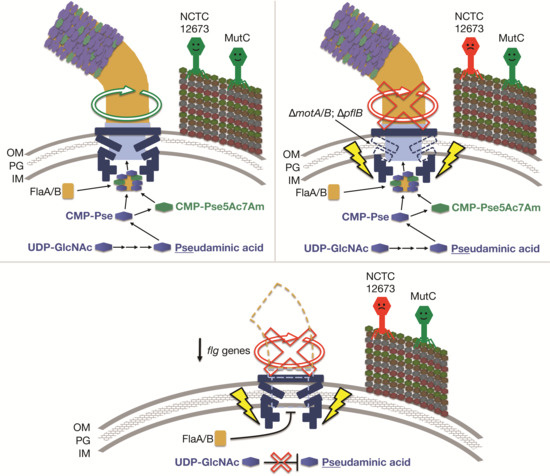
Reduced Infection Efficiency of Phage NCTC 12673 on Non-Motile Campylobacter jejuni Strains Is Related to Oxidative Stress
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Growth Conditions
2.2. Mutagenesis and Complementation
2.3. Phage Propagation and Titration
2.4. Efficiency of Plating (EOP) Assays
2.5. Deoxycholate Assay
2.6. Adsorption Assays
2.7. Total RNA Extraction
2.8. RNA-Sequencing
2.9. Isolation of MutC
2.10. Whole Genome Sequencing and Analysis
3. Results
3.1. Phage NCTC 12673 Requires a Functional Pseudaminic Acid Biosynthetic Pathway for Infection
3.2. Phage NCTC 12673 Adsorbs to Pseudaminic Acid Pathway Mutants at Wild Type Levels
3.3. ΔpseC and ΔpseF Mutants Display No Evidence of Stress Response or Phage Defense, but Downregulate Many Flagellar Genes
3.4. Phage NCTC 12673 Is Unable to Infect Cells in the Absence of the Flagellar Motor Proteins MotA or MotB
3.5. Oxidative Stress Sensitivity of Non-Motile Mutant Strains May Explain Reduced NCTC 12673 Plaquing Efficiency
3.6. A spontaneous NCTC 12673 Mutant Phage, “MutC”, Efficiently Plaques on Both Non-Motile and Oxidative Stress Defense Mutants
3.7. Phage MutC Is Less Impacted by Exposure to the Oxidative Stress-Inducing Agent Deoxycholate Than Phage NCTC 12673
3.8. Genomic Comparison between Phages NCTC 12673 and MutC Predicts Differences in Several Proteins, including FlaGrab, a Flagellar Glycan-Binding Protein
3.9. Differences in Variant Frequency within Two Poly-Adenosine Nucleotide Tracts Suggests MutC Phage Expresses a Truncated FlaGrab Protein More Frequently Than NCTC 12673 Phage
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kaakoush, N.O.; Castaño-Rodríguez, N.; Mitchell, H.M.; Man, S.M. Global epidemiology of Campylobacter infection. Clin. Microbiol. Rev. 2015, 28, 687–720. [Google Scholar] [CrossRef] [Green Version]
- Yuki, N.; Susuki, K.; Koga, M.; Nishimoto, Y.; Odaka, M.; Hirata, K.; Taguchi, K.; Miyatake, T.; Furukawa, K.; Kobata, T.; et al. Carbohydrate mimicry between human ganglioside GM1 and Campylobacter jejuni lipooligosaccharide causes Guillain-Barre syndrome. Proc. Natl. Acad. Sci. USA 2004, 101, 11404–11409. [Google Scholar] [CrossRef] [Green Version]
- Amour, C.; Gratz, J.; Mduma, E.; Svensen, E.; Rogawski, E.T.; McGrath, M.; Seidman, J.C.; McCormick, B.J.J.J.; Shrestha, S.; Samie, A.; et al. Etiology, Risk Factors, and Interactions of Enteric Infections and Malnutrition and the Consequences for Child Health and Development Project (MAL-ED) Network Investigators Epidemiology and Impact of Campylobacter Infection in Children in 8 Low-Resource Settings: Results from the MAL-ED Study. Clin. Infect. Dis. 2016, 63, 1171–1179. [Google Scholar] [CrossRef] [Green Version]
- Hampton, T. Report Reveals Scope of US Antibiotic Resistance Threat. JAMA 2013, 310, 1661. [Google Scholar] [CrossRef]
- Johnson, T.J.; Shank, J.M.; Johnson, J.G. Current and Potential Treatments for Reducing Campylobacter Colonization in Animal Hosts and Disease in Humans. Front. Microbiol. 2017, 8, 487. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wassenaar, T.M. Following an imaginary Campylobacter population from farm to fork and beyond: A bacterial perspective. Lett. Appl. Microbiol. 2011, 53, 253–263. [Google Scholar] [CrossRef] [PubMed]
- Abedon, S.; Kuhl, S.J.; Blasdel, B.G.; Kutter, E.M. Phage treatment of human infections. Bacteriophage 2011, 1, 66–85. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Smet, J.; Hendrix, H.; Blasdel, B.G.; Danis-Wlodarczyk, K.; Lavigne, R. Pseudomonas predators: Understanding and exploiting phage-host interactions. Nat. Rev. Microbiol. 2017, 15, 517–530. [Google Scholar] [CrossRef]
- Connerton, P.L.; Timms, A.R.; Connerton, I.F. Campylobacter bacteriophages and bacteriophage therapy. J. Appl. Microbiol. 2011, 111, 255–265. [Google Scholar] [CrossRef]
- El-Shibiny, A.; Connerton, P.L.; Connerton, I.F. Campylobacter succession in broiler chickens. Vet. Microbiol. 2007, 125, 323–332. [Google Scholar] [CrossRef]
- El-Shibiny, A.; Scott, A.; Timms, A.; Metawea, Y.; Connerton, P.; Connerton, I. Application of a group II Campylobacter bacteriophage to reduce strains of Campylobacter jejuni and Campylobacter coli colonizing broiler chickens. J. Food Prot. 2009, 72, 733–740. [Google Scholar] [CrossRef]
- Karlyshev, A.V.; Ketley, J.M.; Wren, B.W. The Campylobacter jejuni glycome. FEMS Microbiol. Rev. 2005, 29, 377–390. [Google Scholar]
- Nothaft, H.; Szymanski, C.M. Protein glycosylation in bacteria: Sweeter than ever. Nat. Rev. Microbiol. 2010, 8, 765–778. [Google Scholar] [CrossRef] [PubMed]
- Simpson, D.J.; Sacher, J.C.; Szymanski, C.M. Exploring the interactions between bacteriophage-encoded glycan binding proteins and carbohydrates. Curr. Opin. Struct. Biol. 2015, 34, 69–77. [Google Scholar] [CrossRef] [PubMed]
- Sørensen, M.C.H.; Gencay, Y.E.; Birk, T.; Baldvinsson, S.B.; Jäckel, C.; Hammerl, J.A.; Vegge, C.S.; Neve, H.; Brøndsted, L. Primary isolation strain determines both phage type and receptors recognised by Campylobacter jejuni bacteriophages. PLoS ONE 2015, 10, e0116287. [Google Scholar] [CrossRef] [Green Version]
- Coward, C.; Grant, A.J.; Swift, C.; Philp, J.; Towler, R.; Heydarian, M.; Frost, J.A.; Maskell, D.J. Phase-variable surface structures are required for infection of Campylobacter jejuni by bacteriophages. Appl. Environ. Microbiol. 2006, 72, 4638–4647. [Google Scholar] [CrossRef] [Green Version]
- Javed, M.A.; Ackermann, H.W.; Azeredo, J.; Carvalho, C.M.; Connerton, I.; Evoy, S.; Hammerl, J.A.; Hertwig, S.; Lavigne, R.; Singh, A.; et al. A suggested classification for two groups of Campylobacter myoviruses. Arch. Virol. 2014, 159, 181–190. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kropinski, A.M.; Arutyunov, D.; Foss, M.; Cunningham, A.; Ding, W.; Singh, A.; Pavlov, A.R.; Henry, M.; Evoy, S.; Kelly, J.; et al. Genome and proteome of Campylobacter jejuni bacteriophage NCTC 12673. Appl. Environ. Microbiol. 2011, 77, 8265–8271. [Google Scholar] [CrossRef] [Green Version]
- Goon, S.; Kelly, J.F.; Logan, S.M.; Ewing, C.P.; Guerry, P. Pseudaminic acid, the major modification on Campylobacter flagellin, is synthesized via the Cj1293 gene. Mol. Microbiol. 2003, 50, 659–671. [Google Scholar] [CrossRef]
- Guerry, P.; Szymanski, C.M. Campylobacter sugars sticking out. Trends Microbiol. 2008, 16, 428–435. [Google Scholar] [CrossRef]
- Beeby, M.; Ribardo, D.A.; Brennan, C.A.; Ruby, E.G.; Jensen, G.J.; Hendrixson, D.R. Diverse high-torque bacterial flagellar motors assemble wider stator rings using a conserved protein scaffold. Proc. Natl. Acad. Sci. USA 2016, 113, E1917–E1926. [Google Scholar] [CrossRef] [Green Version]
- Flint, A.; Sun, Y.Q.; Butcher, J.; Stahl, M.; Huang, H.; Stintzi, A. Phenotypic screening of a targeted mutant library reveals Campylobacter jejuni defenses against oxidative stress. Infect. Immun. 2014, 82, 2266–2275. [Google Scholar] [CrossRef] [Green Version]
- Sacher, J.C.; Flint, A.; Butcher, J.; Blasdel, B.; Reynolds, H.M.; Lavigne, R.; Stintzi, A.; Szymanski, C.M. Transcriptomic analysis of the Campylobacter jejuni response to T4-like phage NCTC 12673 infection. Viruses 2018, 10, 332. [Google Scholar] [CrossRef] [Green Version]
- Sacher, J.C.; Shajahan, A.; Butcher, J.; Patry, R.T.; Flint, A.; Hendrixson, D.R.; Szymanski, C.M. Binding of phage-encoded FlaGrab to motile Campylobacter jejuni flagella inhibits growth, downregulates energy metabolism, and requires specific flagellar glycans. Front. Microbiol. 2020, 11, 397. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sørensen, M.C.H.; van Alphen, L.B.; Harboe, A.; Li, J.; Christensen, B.B.; Szymanski, C.M.; Brøndsted, L. Bacteriophage F336 recognizes the capsular phosphoramidate modification of Campylobacter jejuni NCTC11168. J. Bacteriol. 2011, 193, 6742–6749. [Google Scholar] [CrossRef] [Green Version]
- Parkhill, J.; Wren, B.W.; Mungall, K.; Ketley, J.M.; Churcher, C.; Basham, D.; Chillingworth, T.; Davies, R.M.; Feltwell, T.; Holroyd, S.; et al. The genome sequence of the food-borne pathogen Campylobacter jejuni reveals hypervariable sequences. Nature 2000, 403, 665–668. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Michael, F.S.; Szymanski, C.M.; Li, J.; Chan, K.H.; Khieu, N.H.; Larocque, S.; Wakarchuk, W.W.; Brisson, J.R.; Monteiro, M.A. The structures of the lipooligosaccharide and capsule polysaccharide of Campylobacter jejuni genome sequenced strain NCTC 11168. Eur. J. Biochem. 2002, 269, 5119–5136. [Google Scholar] [CrossRef] [PubMed]
- Irons, J.; Sacher, J.C.; Szymanski, C.M.; Downs, D.M. Cj1388 is a RidA homolog and is required for flagella biosynthesis and/or function in Campylobacter jejuni. Front. Microbiol. 2019, 10, 2058. [Google Scholar] [CrossRef] [PubMed]
- Javed, M.A.; van Alphen, L.B.; Sacher, J.; Ding, W.; Kelly, J.; Nargang, C.; Smith, D.F.; Cummings, R.D.; Szymanski, C.M. A receptor-binding protein of Campylobacter jejuni bacteriophage NCTC 12673 recognizes flagellin glycosylated with acetamidino-modified pseudaminic acid. Mol. Microbiol. 2015, 95, 101–115. [Google Scholar] [CrossRef] [PubMed]
- Javed, M.A.; Sacher, J.C.; van Alphen, L.B.; Patry, R.T.; Szymanski, C.M. A flagellar glycan-specific protein encoded by Campylobacter phages inhibits host cell growth. Viruses 2015, 7, 6661–6674. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Palyada, K.; Sun, Y.Q.; Flint, A.; Butcher, J.; Naikare, H.; Stintzi, A. Characterization of the oxidative stress stimulon and PerR regulon of Campylobacter jejuni. BMC Genom. 2009, 10, 481. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dwivedi, R.; Nothaft, H.; Garber, J.; Szymanski, C.M. L-Fucose influences chemotaxis and biofilm formation in Campylobacter jejuni. Mol. Microbiol. 2016, 101, 575–589. [Google Scholar] [CrossRef] [Green Version]
- Grajewski, B.A.; Kusek, J.W.; Gelfand, H.M. Development of bacteriophage typing system for Campylobacter jejuni and Campylobacter coli. J. Clin. Microbiol. 1985, 22, 13–18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Frost, J.A.; Kramer, J.M.; Gillanders, S.A. Phage typing of Campylobacter jejuni and Campylobacter coli and its use as an adjunct to serotyping. Epidemiol. Infect. 1999, 123, 47–55. [Google Scholar] [CrossRef]
- Negretti, N.M.; Gourley, C.R.; Clair, G.; Adkins, J.N.; Konkel, M.E. The food-borne pathogen Campylobacter jejuni responds to the bile salt deoxycholate with countermeasures to reactive oxygen species. Sci. Rep. 2017, 7, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Baldvinsson, S.B.; Holst Sørensen, M.C.; Vegge, C.S.; Clokie, M.R.J.; Brøndsted, L. Campylobacter jejuni motility is required for infection of the flagellotropic bacteriophage F341. Appl. Environ. Microbiol. 2014, 80, 7096–7106. [Google Scholar] [CrossRef] [Green Version]
- Palyada, K.; Threadgill, D.; Stintzi, A. Iron acquisition and regulation in Campylobacter jejuni. J. Bacteriol. 2004, 186, 4714–4729. [Google Scholar] [CrossRef] [Green Version]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 1–21. [Google Scholar] [CrossRef] [Green Version]
- Sørensen, M.C.; Gencay, Y.E.; Brøndsted, L. Methods for Initial Characterization of Campylobacter jejuni Bacteriophages. Methods Mol. Biol. 2017, 1512, 91–105. [Google Scholar] [CrossRef]
- McNally, D.J.; Schoenhofen, I.C.; Houliston, R.S.; Khieu, N.H.; Whitfield, D.M.; Logan, S.M.; Brisson, J.R. CMP-Pseudaminic Acid is a Natural Potent Inhibitor of PseB, the First Enzyme of the Pseudaminic Acid Pathway in Campylobacter jejuni and Helicobacter pylori. ChemMedChem Chem. Enabling Drug Discov. 2008, 3, 55–59. [Google Scholar]
- Szymanski, C.M.; Logan, S.M.; Linton, D.; Wren, B.W. Campylobacter—A tale of two protein glycosylation systems. Trends Microbiol. 2003, 11, 233–238. [Google Scholar] [CrossRef]
- Hofreuter, D.; Tsai, J.; Watson, R.O.; Novik, V.; Altman, B.; Benitez, M.; Galán, J.E. Unique features of a highly pathogenic Campylobacter jejuni strain. Infect. Immun. 2006, 74, 4694–4707. [Google Scholar] [CrossRef] [Green Version]
- Logan, S.M. Flagellar glycosylation—A new component of the motility repertoire? Microbiology 2006, 152, 1249–1262. [Google Scholar] [CrossRef] [Green Version]
- Hooton, S.; Connerton, I.F. Campylobacter jejuni acquire new host-derived CRISPR spacers when in association with bacteriophages harboring a CRISPR-like Cas4 protein. Front. Microbiol. 2015, 5, 744. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Radomska, K.A.; Ordoñez, S.R.; Wösten, M.M.; Wagenaar, J.A.; van Putten, J.P. Feedback control of Campylobacter jejuni flagellin levels through reciprocal binding of FliW to flagellin and the global regulator CsrA. Mol. Microbiol. 2016, 102, 207–220. [Google Scholar] [CrossRef]
- Dugar, G.; Svensson, S.L.; Bischler, T.; Wäldchen, S.; Reinhardt, R.; Sauer, M.; Sharma, C.M. The CsrA-FliW network controls polar localization of the dual-function flagellin mRNA in Campylobacter jejuni. Nat. Commun. 2016, 7, 1–18. [Google Scholar] [CrossRef] [Green Version]
- Boll, J.M.; Hendrixson, D.R. A regulatory checkpoint during flagellar biogenesis in Campylobacter jejuni initiates signal transduction to activate transcription of flagellar genes. MBio 2013, 4, e00432-3. [Google Scholar] [CrossRef] [Green Version]
- Gilbreath, J.J.; Cody, W.L.; Merrell, D.S.; Hendrixson, D.R. Change is good: Variations in common biological mechanisms in the epsilonproteobacterial genera Campylobacter and Helicobacter. Microbiol. Mol. Biol. Rev. 2011, 75, 84–132. [Google Scholar] [CrossRef] [Green Version]
- Guerry, P. Campylobacter flagella: Not just for motility. Trends Microbiol. 2007, 15, 456–461. [Google Scholar] [CrossRef]
- Carrillo, C.D.; Taboada, E.; Nash, J.H.; Lanthier, P.; Kelly, J.; Lau, P.C.; Szymanski, C.M. Genome-wide expression analyses of Campylobacter jejuni NCTC11168 reveals coordinate regulation of motility and virulence by flhA. J. Biol. Chem. 2004, 279, 20327–20338. [Google Scholar] [CrossRef] [Green Version]
- Chatterjee, A.; Willett, J.L.; Nguyen, U.T.; Monogue, B.; Palmer, K.L.; Dunny, G.M.; Duerkop, B.A. Parallel genomics uncover novel enterococcal-bacteriophage interactions. MBio 2020, 11, e03120-19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, J.C.; Oh, E.; Kim, J.; Jeon, B. Regulation of oxidative stress resistance in Campylobacter jejuni, a microaerophilic foodborne pathogen. Front. Microbiol. 2016, 6, 751. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Benoit, S.L.; Bayyareddy, K.; Mahawar, M.; Sharp, J.S.; Maier, R.J. Alkyl hydroperoxide reductase repair by Helicobacter pylori methionine sulfoxide reductase. J. Bacteriol. 2015, 195, 5396–5401. [Google Scholar] [CrossRef] [Green Version]
- Benoit, S.L.; Maier, R.J. Helicobacter Catalase Devoid of Catalytic Activity Protects the Bacterium against Oxidative Stress. J. Biol. Chem. 2016, 291, 23366–23373. [Google Scholar] [CrossRef] [Green Version]
- Hofreuter, D.; Novik, V.; Galán, J.E. Metabolic diversity in Campylobacter jejuni enhances specific tissue colonization. Cell Host Microbe 2008, 4, 425–433. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ezraty, B.; Gennaris, A.; Barras, F.; Collet, J.F. Oxidative stress, protein damage and repair in bacteria. Nat. Rev. Microbiol. 2017, 15, 385–396. [Google Scholar] [CrossRef] [PubMed]
- Lango-Scholey, L.; Aidley, J.; Woodacre, A.; Jones, M.A.; Bayliss, C.D. High throughput method for analysis of repeat number for 28 phase variable loci of Campylobacter jejuni strain NCTC11168. PLoS ONE 2016, 11, e0159634. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sørensen, M.C.H.; Vitt, A.; Neve, H.; Soverini, M.; Ahern, S.J.; Klumpp, J.; Brøndsted, L. Campylobacter phages use hypermutable polyG tracts to create phenotypic diversity and evade bacterial resistance. Cell Rep. 2021, 35, 109214. [Google Scholar] [CrossRef]
- Aidley, J.; Holst Sørensen, M.C.; Bayliss, C.D.; Brøndsted, L. Phage exposure causes dynamic shifts in the expression states of specific phase-variable genes of Campylobacter jejuni. Microbiology 2017, 163, 911–919. [Google Scholar] [CrossRef]
- Tormo-Más, M.Á.; Mir, I.; Shrestha, A.; Tallent, S.M.; Campoy, S.; Lasa, Í.; Penadés, J.R. Moonlighting bacteriophage proteins derepress staphylococcal pathogenicity islands. Nature 2010, 465, 779–782. [Google Scholar] [CrossRef] [Green Version]
- Siringan, P.; Connerton, P.L.; Cummings, N.J.; Connerton, I.F. Alternative bacteriophage life cycles: The carrier state of Campylobacter jejuni. Open Biol. 2014, 4, 130200. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sacher, J.C. Insights into the Role of the Flagellar Glycosylation System in Campylobacter jejuni Phage-Host Interactions. Ph.D. Thesis, University of Alberta, Edmonton, AB, Canada, 2018. [Google Scholar]




| Strain | Description/Phenotype | Reference |
|---|---|---|
| C. jejuni NCTC 11168 | Human enteropathy isolate, capsular, motile | [26] |
| C. jejuni NCTC 11168∆motA | Non-motile (paralyzed flagella) | [24] |
| C. jejuni NCTC 11168∆motB | Non-motile (paralyzed flagella) | [24] |
| C. jejuni NCTC 11168∆kpsM | Acapsular | [27] |
| C. jejuni NCTC 11168∆pseC | Non-motile (aflagellate) | [28] |
| C. jejuni NCTC 11168∆pseH | Non-motile (aflagellate) | [29] |
| C. jejuni NCTC 11168∆pseH/+pseH | Motile | This work |
| C. jejuni NCTC 11168∆pseG | Non-motile (aflagellate) | [30] |
| C. jejuni NCTC 11168∆pseF | Non-motile (aflagellate) | This work |
| C. jejuni NCTC 11168∆katA | Hypersensitive to oxidative stress (lacks catalase) | [31] |
| C. jejuni NCTC 11168∆ahpC | Hypersensitive to oxidative stress (lacks alkyl hydroxyperoxide reductase) | [31] |
| C. jejuni NCTC 11168∆sodB | Hypersensitive to oxidative stress (lacks superoxide dismutase) | [31] |
| C. jejuni NCTC 11168∆flaA | Non-motile (aflagellate) | [32] |
| NCTC 12673 | UK phage typing scheme phage 1 | [18,33] |
| MutC | Spontaneous variant of NCTC 12673 | This work |
| Gene. | Predicted Function | Variant | Phage | Position within Genome | Nucleotide Change | Amino Acid Change | Variant Frequency |
|---|---|---|---|---|---|---|---|
| gp041 | Gp6 baseplate wedge subunit | 1 | MutC | 31,844 | A → G | L → S | 72.70% |
| 2 | MutC | 31,885 | C → T | M → I | 25.60% | ||
| gp047/flagrab | FlaGrab, flagellar glycan binding protein | 1 | MutC | 40,821 | (A)7 → (A)6 | Frame Shift | 68.80% |
| 2 | NCTC 12673 | 40,935 | (A)7 → (A)6 | Frame Shift | 33.20% | ||
| MutC | 40,935 | (A)7 → (A)6 | Frame Shift | 25.30% | |||
| gp058 | Hef59 homing endonuclease | 1 | MutC | 50,606 | AA → GG | E → G | 69.8% |
| gp114/gp115 | Hypothetical protein | 1 | MutC | 100,737 | (CC)4 → (CC)5 | Frame Shift | 37.00% |
| NCTC 12673 | 100,737 | (C)9 → (C)10 | Frame Shift | 31.20% | |||
| MutC | 100,737 | (C)9 → (C)10 | Frame Shift | 43.70% | |||
| gp116 | Hypothetical protein | 1 | NCTC 12673 | 101,692 | (C)11 → (C)12 | Frame Shift | 28.90% |
| 2 | NCTC 12673 | 101,691 | (C)11 → (C)10 | Frame Shift | 27.10% | ||
| MutC | 101,691 | (C)11 → (C)10 | Frame Shift | 62.80% | |||
| 3 | MutC | 101,202 | G → A | T → M | 68.70% | ||
| 4 | NCTC 12673 | 101,112 | G → T | T → N | 73.80% | ||
| gp167 | Hef168 homing endonuclease | 1 | MutC | 132,316 | C → T | D → N | 64.00% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sacher, J.C.; Javed, M.A.; Crippen, C.S.; Butcher, J.; Flint, A.; Stintzi, A.; Szymanski, C.M. Reduced Infection Efficiency of Phage NCTC 12673 on Non-Motile Campylobacter jejuni Strains Is Related to Oxidative Stress. Viruses 2021, 13, 1955. https://doi.org/10.3390/v13101955
Sacher JC, Javed MA, Crippen CS, Butcher J, Flint A, Stintzi A, Szymanski CM. Reduced Infection Efficiency of Phage NCTC 12673 on Non-Motile Campylobacter jejuni Strains Is Related to Oxidative Stress. Viruses. 2021; 13(10):1955. https://doi.org/10.3390/v13101955
Chicago/Turabian StyleSacher, Jessica C., Muhammad Afzal Javed, Clay S. Crippen, James Butcher, Annika Flint, Alain Stintzi, and Christine M. Szymanski. 2021. "Reduced Infection Efficiency of Phage NCTC 12673 on Non-Motile Campylobacter jejuni Strains Is Related to Oxidative Stress" Viruses 13, no. 10: 1955. https://doi.org/10.3390/v13101955
APA StyleSacher, J. C., Javed, M. A., Crippen, C. S., Butcher, J., Flint, A., Stintzi, A., & Szymanski, C. M. (2021). Reduced Infection Efficiency of Phage NCTC 12673 on Non-Motile Campylobacter jejuni Strains Is Related to Oxidative Stress. Viruses, 13(10), 1955. https://doi.org/10.3390/v13101955