Nucleocytoplasmic Trafficking Perturbation Induced by Picornaviruses
Abstract
:1. Introduction: Nucleocytoplasmic Trafficking of Proteins and Picornavirus Replication
2. Picornaviruses Trigger the Mislocalization of Host Proteins in Infected Cells
2.1. Which Trafficking Pathways Are Affected?
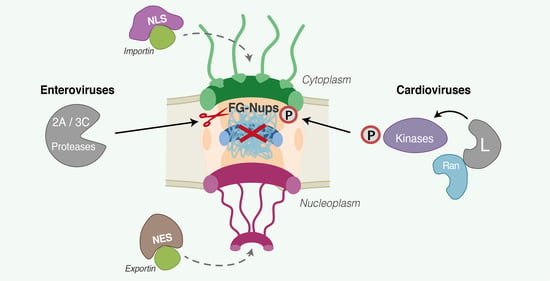
2.2. FG-Nucleoporins as the Main Targets for Nucleocytoplasmic Disturbance
2.3. Different Picornaviruses Acting with Different Proteins
| Nucleoporins | FG-Repeat | Enterovirus | Cardiovirus | ||||
|---|---|---|---|---|---|---|---|
| Poliovirus | Enterovirus 70 & 71 | Coxsackievirus B3 | Rhinovirus | EMCV | TMEV | ||
| NUP35 | + | C [45] | NT | NT | NT | NT | NT |
| NUP54 | + | C [45] | NT | C * | NT | NT | NT |
| NUP58 | + | C [45] | NT | NT | NT | NT | NT |
| NUP62 | + | C [26,39,44,45] (2Apro [40]) | NT | C [43] | C [26,53] (2Apro [38,46,47,51], 3Cpro [46]) | P [56,57] | P * |
| NUP98 | + | C [44,45,55] (2Apro [40,47]) | C [55] | C (2Apro [52,55]) | C [53,55] (2Apro [44,47,51]) | NT | P [41] |
| NUP153 | + | C [26,39,44,45] (2Apro [40]) | NT | C [43] | C [26] (2Apro [38,51], 3Cpro [53,54]) | P [56,57] | P * |
| NUP214 | + | C [45] | NT | C * | C (3Cpro [54]) | P [56,57] | P * |
| NUP358 | + | C [45] | NT | NT | C (3Cpro [54]) | - [56] | NT |
| NLP1 | + | C [45] | NT | NT | NT | NT | NT |
| POM121 | + | C [45] | NT | NT | NT | NT | P * |
| TPR | - | C [45] | NT | NT | NT | NT | NT |
| RAE1 | - | C * | NT | C * | NT | NT | NT |
2.4. Are Picornaviruses Dismantling the NPC Structure?
2.5. RAN and Karyopherins Are among the Targets in the Soluble Phase
3. Conclusions and Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Krull, S.; Thyberg, J.; Björkroth, B.; Rackwitz, H.-R.; Cordes, V.C. Nucleoporins as Components of the Nuclear Pore Complex Core Structure and Tpr as the Architectural Element of the Nuclear Basket. Mol. Biol. Cell 2004, 15, 4261–4277. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rout, M.P.; Aitchison, J.D.; Suprapto, A.; Hjertaas, K.; Zhao, Y.; Chait, B.T. The yeast nuclear pore complex: Composition, architecture, and transport mechanism. J. Cell Biol. 2000, 148, 635–651. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schwartz, T.U. The Structure Inventory of the Nuclear Pore Complex. J. Mol. Biol. 2016, 428, 1986–2000. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lim, R.Y.H.; Fahrenkrog, B.; Köser, J.; Schwarz-Herion, K.; Deng, J.; Aebi, U. Nanomechanical Basis of Selective Gating by the Nuclear Pore Complex. Science 2007, 318, 640–643. [Google Scholar] [CrossRef]
- Frey, S.; Richter, R.P.; Görlich, D. FG-Rich Repeats of Nuclear Pore Proteins Form a Three-Dimensional Meshwork with Hydrogel-Like Properties. Science 2006, 314, 815–817. [Google Scholar] [CrossRef] [Green Version]
- Rexach, M.; Blobel, G. Protein import into nuclei: Association and dissociation reactions involving transport substrate, transport factors, and nucleoporins. Cell 1995, 83, 683–692. [Google Scholar] [CrossRef] [Green Version]
- Beck, M.; Hurt, E. The nuclear pore complex: Understanding its function through structural insight. Nat. Rev. Mol. Cell Biol. 2017, 18, 73–89. [Google Scholar] [CrossRef]
- Hampoelz, B.; Andres-Pons, A.; Kastritis, P.; Beck, M. Structure and Assembly of the Nuclear Pore Complex. Annu. Rev. Biophys. 2019, 48, 515–536. [Google Scholar] [CrossRef]
- Zell, R.; Delwart, E.; Gorbalenya, A.; Hovi, T.; King, A.M.Q.; Knowles, N.J.; Lindberg, A.M.; Pallansch, M.A.; Palmenberg, A.C.; Reuter, G.; et al. ICTV Virus Taxonomy Profile: Picornaviridae. J. Gen. Virol. 2017, 98, 2421–2422. [Google Scholar] [CrossRef]
- Pelletier, J.; Sonenberg, N. Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature 1988, 334, 320–325. [Google Scholar] [CrossRef]
- Jang, S.K.; Kräusslich, H.G.; Nicklin, M.J.; Duke, G.M.; Palmenberg, A.C.; Wimmer, E. A segment of the 5’ nontranslated region of encephalomyocarditis virus RNA directs internal entry of ribosomes during in vitro translation. J. Virol. 1988, 62, 2636–2643. [Google Scholar] [CrossRef] [Green Version]
- Lee, K.-M.; Chen, C.-J.; Shih, S.-R. Regulation Mechanisms of Viral IRES-Driven Translation. Trends Microbiol. 2017, 25, 546–561. [Google Scholar] [CrossRef]
- Martinez-Salas, E.; Francisco-Velilla, R.; Fernandez, J.; Embarek, A.M. Insights into Structural and Mechanistic Features of Viral IRES Elements. Front. Microbiol. 2018, 8, 2629. [Google Scholar] [CrossRef] [Green Version]
- Palmenberg, A.C. Proteolytic processing of picornaviral polyprotein. Annu. Rev. Microbiol. 1990, 44, 603–623. [Google Scholar] [CrossRef]
- Donnelly, M.L.; Hughes, L.E.; Luke, G.; Mendoza, H.; Ten Dam, E.; Gani, D.; Ryan, M.D. The ‘cleavage’ activities of foot-and-mouth disease virus 2A site-directed mutants and naturally occurring ‘2A-like’ sequences. J. Gen. Virol. 2001, 82, 1027–1041. [Google Scholar] [CrossRef]
- Donnelly, M.L.; Luke, G.; Mehrotra, A.; Li, X.; Hughes, L.E.; Gani, D.; Ryan, M.D. Analysis of the aphthovirus 2A/2B polyprotein ‘cleavage’ mechanism indicates not a proteolytic reaction, but a novel translational effect: A putative ribosomal ‘skip’. J. Gen. Virol. 2001, 82, 1013–1025. [Google Scholar] [CrossRef]
- Jesús-González, D.; Adrián, L.; Palacios-Rápalo, S.; Reyes-Ruiz, J.M.; Osuna-Ramos, J.F.; Cordero-Rivera, C.D.; Farfan-Morales, C.N.; Gutiérrez-Escolano, A.L.; del Ángel, R.M. The Nuclear Pore Complex Is a Key Target of Viral Proteases to Promote Viral Replication. Viruses 2021, 13, 706. [Google Scholar] [CrossRef]
- Gustin, K.E. Inhibition of nucleo-cytoplasmic trafficking by RNA viruses: Targeting the nuclear pore complex. Virus Res. 2003, 95, 35–44. [Google Scholar] [CrossRef]
- Lloyd, R.E. Nuclear proteins hijacked by mammalian cytoplasmic plus strand RNA viruses. Virology 2015, 479-480, 457–474. [Google Scholar] [CrossRef] [Green Version]
- Pollack, R.; Goldman, R. Synthesis of Infective Poliovirus in BSC-1 Monkey Cells Enucleated with Cytochalasin B. Science 1973, 179, 915–916. [Google Scholar] [CrossRef]
- Follett, E.A.C.; Pringle, C.R.; Pennington, T.H. Virus Development in Enucleate Cells: Echovirus, Poliovirus, Pseudorabies Virus, Reovirus, Respiratory Syncytial Virus and Semliki Forest Virus. J. Gen. Virol. 1975, 26, 183–196. [Google Scholar] [CrossRef] [PubMed]
- Molla, A.; Paul, A.V.; Wimmer, E. Cell-free, de novo synthesis of poliovirus. Science 1991, 254, 1647–1651. [Google Scholar] [CrossRef]
- Svitkin, Y.V.; Sonenberg, N. Cell-Free Synthesis of Encephalomyocarditis Virus. J. Virol. 2003, 77, 6551–6555. [Google Scholar] [CrossRef] [Green Version]
- Barton, D.J.; Flanegan, J.B. Coupled translation and replication of poliovirus RNA in vitro: Synthesis of functional 3D polymerase and infectious virus. J. Virol. 1993, 67, 822–831. [Google Scholar] [CrossRef] [Green Version]
- McBride, A.E.; Schlegel, A.; Kirkegaard, K. Human protein Sam68 relocalization and interaction with poliovirus RNA polymerase in infected cells. Proc. Natl. Acad. Sci. USA 1996, 93, 2296–2301. [Google Scholar] [CrossRef] [Green Version]
- Gustin, K.E.; Sarnow, P. Inhibition of Nuclear Import and Alteration of Nuclear Pore Complex Composition by Rhinovirus. J. Virol. 2002, 76, 8787–8796. [Google Scholar] [CrossRef] [Green Version]
- Waggoner, S.; Sarnow, P. Viral ribonucleoprotein complex formation and nucleolar-cytoplasmic relocalization of nucleolin in poliovirus-infected cells. J. Virol. 1998, 72, 6699–6709. [Google Scholar] [CrossRef] [Green Version]
- Meerovitch, K.; Svitkin, Y.V.; Lee, H.S.; Lejbkowicz, F.; Kenan, D.J.; Chan, E.; Agol, V.I.; Keene, J.D.; Sonenberg, N. La autoantigen enhances and corrects aberrant translation of poliovirus RNA in reticulocyte lysate. J. Virol. 1993, 67, 3798–3807. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Borman, A.; Howell, M.T.; Patton, J.G.; Jackson, R.J. The involvement of a spliceosome component in internal initiation of human rhinovirus RNA translation. J. Gen. Virol. 1993, 74, 1775–1788. [Google Scholar] [CrossRef] [PubMed]
- Hellen, C.U.; Witherell, G.W.; Schmid, M.; Shin, S.H.; Pestova, T.V.; Gil, A.; Wimmer, E. A cytoplasmic 57-kDa protein that is required for translation of picornavirus RNA by internal ribosomal entry is identical to the nuclear pyrimidine tract-binding protein. Proc. Natl. Acad. Sci. USA 1993, 90, 7642–7646. [Google Scholar] [CrossRef] [Green Version]
- Delhaye, S.; Van Pesch, V.; Michiels, T. The Leader Protein of Theiler’s Virus Interferes with Nucleocytoplasmic Trafficking of Cellular Proteins. J. Virol. 2004, 78, 4357–4362. [Google Scholar] [CrossRef] [Green Version]
- Belov, G.A.; Evstafieva, A.G.; Rubtsov, Y.P.; Mikitas, O.V.; Vartapetian, A.B.; Agol, V.I. Early Alteration of Nucleocytoplasmic Traffic Induced by Some RNA Viruses. Virology 2000, 275, 244–248. [Google Scholar] [CrossRef] [Green Version]
- Kalderon, D.; Roberts, B.L.; Richardson, W.D.; Smith, A.E. A short amino acid sequence able to specify nuclear location. Cell 1984, 39, 499–509. [Google Scholar] [CrossRef]
- Pollard, V.W.; Michael, W.; Nakielny, S.; Siomi, M.C.; Wang, F.; Dreyfuss, G. A Novel Receptor-Mediated Nuclear Protein Import Pathway. Cell 1996, 86, 985–994. [Google Scholar] [CrossRef] [Green Version]
- Kataoka, N.; Bachorik, J.L.; Dreyfuss, G. Transportin-SR, a Nuclear Import Receptor for SR Proteins. J. Cell Biol. 1999, 145, 1145–1152. [Google Scholar] [CrossRef] [Green Version]
- Wen, W.; Meinkotht, J.L.; Tsien, R.Y.; Taylor, S.S. Identification of a signal for rapid export of proteins from the nucleus. Cell 1995, 82, 463–473. [Google Scholar] [CrossRef] [Green Version]
- Ciomperlik, J.J.; Basta, H.A.; Palmenberg, A.C. Three cardiovirus Leader proteins equivalently inhibit four different nucleocytoplasmic trafficking pathways. Virology 2015, 484, 194–202. [Google Scholar] [CrossRef] [Green Version]
- Watters, K.; Inankur, B.; Gardiner, J.; Warrick, J.; Sherer, N.M.; Yin, J.; Palmenberg, A.C. Differential Disruption of Nucleocytoplasmic Trafficking Pathways by Rhinovirus 2A Proteases. J. Virol. 2017, 91, e02472-16. [Google Scholar] [CrossRef] [Green Version]
- Gustin, K.E.; Sarnow, P. Effects of poliovirus infection on nucleo-cytoplasmic trafficking and nuclear pore complex composition. Embo. J. 2001, 20, 240–249. [Google Scholar] [CrossRef] [Green Version]
- Castelló, A.; Izquierdo, J.M.; Welnowska, E.; Carrasco, L. RNA nuclear export is blocked by poliovirus 2A protease and is concomitant with nucleoporin cleavage. J. Cell Sci. 2009, 122, 3799–3809. [Google Scholar] [CrossRef] [Green Version]
- Ricour, C.; Delhaye, S.; Hato, S.V.; Olenyik, T.D.; Michel, B.; Van Kuppeveld, F.J.M.; Gustin, K.E.; Michiels, T. Inhibition of mRNA export and dimerization of interferon regulatory factor 3 by Theiler’s virus leader protein. J. Gen. Virol. 2009, 90, 177–186. [Google Scholar] [CrossRef]
- Porter, F.W.; Bochkov, Y.; Albee, A.J.; Wiese, C.; Palmenberg, A.C. A picornavirus protein interacts with Ran-GTPase and disrupts nucleocytoplasmic transport. Proc. Natl. Acad. Sci. USA 2006, 103, 12417–12422. [Google Scholar] [CrossRef] [Green Version]
- Lidsky, P.V.; Hato, S.; Bardina, M.V.; Aminev, A.G.; Palmenberg, A.C.; Sheval, E.V.; Polyakov, V.Y.; Van Kuppeveld, F.J.M.; Agol, V.I. Nucleocytoplasmic Traffic Disorder Induced by Cardioviruses. J. Virol. 2006, 80, 2705–2717. [Google Scholar] [CrossRef] [Green Version]
- Park, N.; Katikaneni, P.; Skern, T.; Gustin, K.E. Differential Targeting of Nuclear Pore Complex Proteins in Poliovirus-Infected Cells. J. Virol. 2008, 82, 1647–1655. [Google Scholar] [CrossRef] [Green Version]
- Krull, S.; Dörries, J.; Boysen, B.; Reidenbach, S.; Magnius, L.; Norder, H.; Thyberg, J.; Cordes, V.C. Protein Tpr is required for establishing nuclear pore-associated zones of heterochromatin exclusion. Embo. J. 2010, 29, 1659–1673. [Google Scholar] [CrossRef] [Green Version]
- Park, N.; Skern, T.; Gustin, K.E. Specific Cleavage of the Nuclear Pore Complex Protein Nup62 by a Viral Protease. J. Biol. Chem. 2010, 285, 28796–28805. [Google Scholar] [CrossRef] [Green Version]
- Park, N.; Schweers, N.J.; Gustin, K.E. Selective Removal of FG Repeat Domains from the Nuclear Pore Complex by Enterovirus 2A(pro). J. Virol. 2015, 89, 11069–11079. [Google Scholar] [CrossRef] [Green Version]
- Hülsmann, B.B.; Labokha, A.A.; Görlich, D. The Permeability of Reconstituted Nuclear Pores Provides Direct Evidence for the Selective Phase Model. Cell 2012, 150, 738–751. [Google Scholar] [CrossRef] [Green Version]
- Patel, S.S.; Belmont, B.; Sante, J.M.; Rexach, M.F. Natively Unfolded Nucleoporins Gate Protein Diffusion across the Nuclear Pore Complex. Cell 2007, 129, 83–96. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Belov, G.A.; Lidsky, P.V.; Mikitas, O.V.; Egger, D.; Lukyanov, K.A.; Bienz, K.; Agol, V.I. Bidirectional increase in permeability of nuclear envelope upon poliovirus infection and accompanying alterations of nuclear pores. J. Virol. 2004, 78, 10166–10177. [Google Scholar] [CrossRef] [Green Version]
- Watters, K.; Palmenberg, A.C. Differential Processing of Nuclear Pore Complex Proteins by Rhinovirus 2A Proteases from Different Species and Serotypes. J. Virol. 2011, 85, 10874–10883. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hanson, P.J.; Hossain, A.R.; Qiu, Y.; Zhang, H.M.; Zhao, G.; Li, C.; Lin, V.; Sulaimon, S.; Vlok, M.; Fung, G.; et al. Cleavage and Sub-Cellular Redistribution of Nuclear Pore Protein 98 by Coxsackievirus B3 Protease 2A Impairs Cardioprotection. Front. Cell. Infect. Microbiol. 2019, 9, 265. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Walker, E.J.; Younessi, P.; Fulcher, A.J.; McCuaig, R.; Thomas, B.J.; Bardin, P.G.; Jans, D.A.; Ghildyal, R. Rhinovirus 3C Protease Facilitates Specific Nucleoporin Cleavage and Mislocalisation of Nuclear Proteins in Infected Host Cells. PLoS ONE 2013, 8, e71316. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ghildyal, R.; Jordan, B.; Li, D.; Dagher, H.; Bardin, P.G.; Gern, J.E.; Jans, D.A. Rhinovirus 3C protease can localize in the nucleus and alter active and passive nucleocytoplasmic transport. J. Virol. 2009, 83, 7349–7352. [Google Scholar] [CrossRef] [Green Version]
- Saeed, M.; Kapell, S.; Hertz, N.T.; Wu, X.; Bell, K.; Ashbrook, A.W.; Mark, M.T.; Zebroski, H.A.; Neal, M.L.; Flodström-Tullberg, M.; et al. Defining the proteolytic landscape during enterovirus infection. PLoS Pathog. 2020, 16, e1008927. [Google Scholar] [CrossRef]
- Porter, F.W.; Palmenberg, A.C. Leader-induced phosphorylation of nucleoporins correlates with nuclear trafficking inhibition by cardioviruses. J. Virol. 2009, 83, 1941–1951. [Google Scholar] [CrossRef] [Green Version]
- Porter, F.W.; Brown, B.; Palmenberg, A.C. Nucleoporin Phosphorylation Triggered by the Encephalomyocarditis Virus Leader Protein Is Mediated by Mitogen-Activated Protein Kinases. J. Virol. 2010, 84, 12538–12548. [Google Scholar] [CrossRef] [Green Version]
- Freundt, E.C.; Drappier, M.; Michiels, T. Innate Immune Detection of Cardioviruses and Viral Disruption of Interferon Signaling. Front. Microbiol. 2018, 9, 2448. [Google Scholar] [CrossRef] [Green Version]
- Brahic, M.; Bureau, J.-F.; Michiels, T. The genetics of the persistent infection and demyelinating disease caused by theiler’s virus. Annu. Rev. Microbiol. 2005, 59, 279–298. [Google Scholar] [CrossRef]
- Basta, H.; Bacot-Davis, V.R.; Ciomperlik, J.J.; Palmenberg, A.C. Encephalomyocarditis Virus Leader Is Phosphorylated by CK2 and Syk as a Requirement for Subsequent Phosphorylation of Cellular Nucleoporins. J. Virol. 2014, 88, 2219–2226. [Google Scholar] [CrossRef] [Green Version]
- Basta, H.A.; Palmenberg, A.C. AMP-activated protein kinase phosphorylates EMCV, TMEV and SafV leader proteins at different sites. Virology 2014, 462-463, 236–240. [Google Scholar] [CrossRef] [Green Version]
- Bardina, M.V.; Lidsky, P.V.; Sheval, E.V.; Fominykh, K.V.; van Kuppeveld, F.J.M.; Polyakov, V.Y.; Agol, V.I. Mengovirus-Induced Rearrangement of the Nuclear Pore Complex: Hijacking Cellular Phosphorylation Machinery. J. Virol. 2009, 83, 3150–3161. [Google Scholar] [CrossRef] [Green Version]
- Bacot-Davis, V.R.; Palmenberg, A.C. Encephalomyocarditis virus Leader protein hinge domain is responsible for interactions with Ran GTPase. Virology 2013, 443, 177–185. [Google Scholar] [CrossRef]
- Bacot-Davis, V.R.; Ciomperlik, J.J.; Basta, H.A.; Cornilescu, C.C.; Palmenberg, A.C. Solution structures of Mengovirus Leader protein, its phosphorylated derivatives, and in complex with nuclear transport regulatory protein, RanGTPase. Proc. Natl. Acad. Sci. USA 2014, 111, 15792–15797. [Google Scholar] [CrossRef] [Green Version]
- Petty, R.V.; Palmenberg, A.C. Guanine-Nucleotide Exchange Factor RCC1 Facilitates a Tight Binding between the Encephalomyocarditis Virus Leader and Cellular Ran GTPase. J. Virol. 2013, 87, 6517–6520. [Google Scholar] [CrossRef] [Green Version]
- Ciomperlik, J.J.; Basta, H.A.; Palmenberg, A.C. Cardiovirus Leader proteins bind exportins: Implications for virus replication and nucleocytoplasmic trafficking inhibition. Virology 2016, 487, 19–26. [Google Scholar] [CrossRef] [Green Version]
- Du, Y.; Bi, J.; Liu, J.; Liu, X.; Wu, X.; Jiang, P.; Yoo, D.; Zhang, Y.; Wu, J.; Wan, R.; et al. 3Cpro of Foot-and-Mouth Disease Virus Antagonizes the Interferon Signaling Pathway by Blocking STAT1/STAT2 Nuclear Translocation. J. Virol. 2014, 88, 4908–4920. [Google Scholar] [CrossRef] [Green Version]
- Wang, C.; Sun, M.; Yuan, X.; Ji, L.; Jin, Y.; Cardona, C.J.; Xing, Z. Enterovirus 71 suppresses interferon responses by blocking Janus kinase (JAK)/signal transducer and activator of transcription (STAT) signaling through inducing karyopherin-α1 degradation. J. Biol. Chem. 2017, 292, 10262–10274. [Google Scholar] [CrossRef] [Green Version]
- Peng, N.; Yang, X.; Zhu, C.; Zhou, L.; Yu, H.; Li, M.; Lin, Y.; Wang, X.; Li, Q.; She, Y.; et al. MicroRNA-302 Cluster Downregulates Enterovirus 71–Induced Innate Immune Response by Targeting KPNA2. J. Immunol. 2018, 201, 145–156. [Google Scholar] [CrossRef] [Green Version]
- Meerovitch, K.; Pelletier, J.; Sonenberg, N. A cellular protein that binds to the 5’-noncoding region of poliovirus RNA: Implications for internal translation initiation. Genes Dev. 1989, 3, 1026–1034. [Google Scholar] [CrossRef] [Green Version]
- Costa-Mattioli, M.; Svitkin, Y.; Sonenberg, N. La Autoantigen Is Necessary for Optimal Function of the Poliovirus and Hepatitis C Virus Internal Ribosome Entry Site In Vivo and In Vitro. Mol. Cell. Biol. 2004, 24, 6861–6870. [Google Scholar] [CrossRef] [Green Version]
- Shiroki, K.; Isoyama, T.; Kuge, S.; Ishii, T.; Ohmi, S.; Hata, S.; Suzuki, K.; Takasaki, Y.; Nomoto, A. Intracellular redistribution of truncated La protein produced by poliovirus 3Cpro-mediated cleavage. J. Virol. 1999, 73, 2193–2200. [Google Scholar] [CrossRef] [Green Version]
- Hato, S.V.; Ricour, C.; Schulte, B.M.; Lanke, K.H.W.; De Bruijni, M.; Zoll, J.; Melchers, W.; Michiels, T.; Van Kuppeveld, F.J.M. The mengovirus leader protein blocks interferon-α/β gene transcription and inhibits activation of interferon regulatory factor 3. Cell. Microbiol. 2007, 9, 2921–2930. [Google Scholar] [CrossRef]
- Mettenleiter, T.C. Breaching the Barrier—The Nuclear Envelope in Virus Infection. J. Mol. Biol. 2016, 428, 1949–1961. [Google Scholar] [CrossRef]
- De Jesús-González, L.A.; Cervantes-Salazar, M.; Reyes-Ruiz, J.M.; Osuna-Ramos, J.F.; Farfán-Morales, C.N.; Palacios-Rápalo, S.N.; Pérez-Olais, J.H.; Cordero-Rivera, C.D.; Hurtado-Monzón, A.M.; Ruíz-Jiménez, F.; et al. The Nuclear Pore Complex: A Target for NS3 Protease of Dengue and Zika Viruses. Viruses 2020, 12, 583. [Google Scholar] [CrossRef]
- Kato, K.; Ikliptikawati, D.K.; Kobayashi, A.; Kondo, H.; Lim, K.; Hazawa, M.; Wong, R.W. Overexpression of SARS-CoV-2 protein ORF6 dislocates RAE1 and NUP98 from the nuclear pore complex. Biochem. Biophys. Res. Commun. 2021, 536, 59–66. [Google Scholar] [CrossRef]
- Addetia, A.; Lieberman, N.A.; Phung, Q.; Hsiang, T.Y.; Xie, H.; Roychoudhury, P.; Shrestha, L.; Loprieno, M.A.; Huang, M.L.; Gale, M., Jr.; et al. SARS-CoV-2 ORF6 Disrupts Bidirectional Nucleocytoplasmic Transport through Interactions with Rae1 and Nup98. mBio 2021, 12, e00065-21. [Google Scholar] [CrossRef]
- Miorin, L.; Kehrer, T.; Sanchez-Aparicio, M.T.; Zhang, K.; Cohen, P.; Patel, R.S.; Cupic, A.; Makio, T.; Mei, M.; Moreno, E.; et al. SARS-CoV-2 Orf6 hijacks Nup98 to block STAT nuclear import and antagonize interferon signaling. Proc. Natl. Acad. Sci. USA 2020, 117, 28344–28354. [Google Scholar] [CrossRef]
- Rolhion, N.; Furniss, R.C.D.; Grabe, G.; Ryan, A.; Liu, M.; Matthews, S.A.; Holden, D.W. Inhibition of Nuclear Transport of NF-ĸB p65 by the Salmonella Type III Secretion System Effector SpvD. PLoS Pathog. 2016, 12, e1005653. [Google Scholar] [CrossRef] [PubMed]
- Evans, S.M.; Rodino, K.G.; Adcox, H.E.; Carlyon, J.A. Orientia tsutsugamushi uses two Ank effectors to modulate NF-κB p65 nuclear transport and inhibit NF-κB transcriptional activation. PLoS Pathog. 2018, 14, e1007023. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Burette, M.; Allombert, J.; Lambou, K.; Maarifi, G.; Nisole, S.; Case, E.D.R.; Blanchet, F.P.; Hassen-Khodja, C.; Cabantous, S.; Samuel, J.; et al. Modulation of innate immune signaling by a Coxiella burnetii eukaryotic-like effector protein. Proc. Natl. Acad. Sci. USA 2020, 117, 13708–13718. [Google Scholar] [CrossRef] [PubMed]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lizcano-Perret, B.; Michiels, T. Nucleocytoplasmic Trafficking Perturbation Induced by Picornaviruses. Viruses 2021, 13, 1210. https://doi.org/10.3390/v13071210
Lizcano-Perret B, Michiels T. Nucleocytoplasmic Trafficking Perturbation Induced by Picornaviruses. Viruses. 2021; 13(7):1210. https://doi.org/10.3390/v13071210
Chicago/Turabian StyleLizcano-Perret, Belén, and Thomas Michiels. 2021. "Nucleocytoplasmic Trafficking Perturbation Induced by Picornaviruses" Viruses 13, no. 7: 1210. https://doi.org/10.3390/v13071210
APA StyleLizcano-Perret, B., & Michiels, T. (2021). Nucleocytoplasmic Trafficking Perturbation Induced by Picornaviruses. Viruses, 13(7), 1210. https://doi.org/10.3390/v13071210