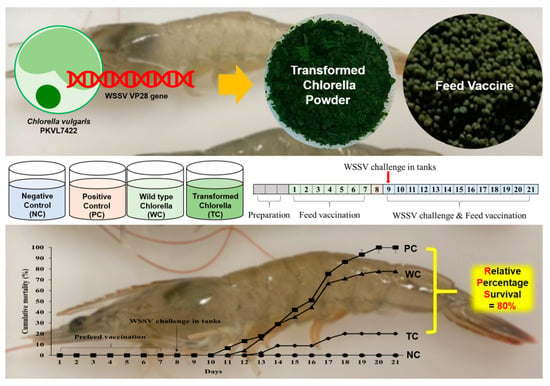
Efficacy of White Spot Syndrome Virus Protein VP28-Expressing Chlorella vulgaris as an Oral Vaccine for Shrimp
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chlorella vulgaris (C. vulgaris) PKVL7422 Growth Conditions
2.2. Construction of the VP28 Expression Vector
2.3. C. vulgaris Transformation
2.4. DNA Extraction and PCR Confirmation
2.5. Confirmation of VP28 Expression in Transformed C. vulgaris
2.6. Shrimp
2.7. Immunization of L. vannamei
2.8. WSSV Challenge
2.9. Reverse-Transcription qPCR (RT-qPCR) Analysis of Immune-Related Genes
2.10. Statistical Analyses
3. Results
3.1. Selection of Transformed Chlorella vulgaris (C. vulgaris)
3.2. PCR Analysis of the Inoculum and WSSV-Challenged Shrimp
3.3. Efficacy of the Oral Vaccine
3.4. RT-qPCR Analysis of Immune Response Genes
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Tacon, A.G. Trends in global aquaculture and aquafeed production: 2000–2017. Rev. Fish. Sci. Aquac. 2020, 28, 43–56. [Google Scholar] [CrossRef]
- FAO. The State of World Fisheries and Aquaculture 2018—Meeting the Sustainable Development Goals; FAO: Rome, Italy, 2018. [Google Scholar]
- Patil, P.K.; Geetha, R.; Ravisankar, T.; Avunje, S.; Solanki, H.G.; Abraham, T.J.; Vinoth, S.P.; Jithendran, K.P.; Alavandi, S.V.; Vijayan, K.K. Economic loss due to diseases in Indian shrimp farming with special reference to Enterocytozoon hepatopenaei (EHP) and white spot syndrome virus (WSSV). Aquaculture 2021, 533, 736231. [Google Scholar] [CrossRef]
- Thornber, K.; Verner-Jeffreys, D.; Hinchliffe, S.; Rahman, M.M.; Bass, D.; Tyler, C.R. Evaluating antimicrobial resistance in the global shrimp industry. Rev. Aquac. 2020, 12, 966–986. [Google Scholar] [CrossRef] [PubMed]
- Amar, E.C.; Faisan, J.P., Jr.; Gapasin, R.S. Field efficacy evaluation of a formalin-inactivated white spot syndrome virus (WSSV) vaccine for the preventive management of WSSV infection in shrimp grow-out ponds. Aquaculture 2021, 531, 735907. [Google Scholar] [CrossRef]
- Kumar, R.; Huang, J.Y.; Ng, Y.S.; Chen, C.Y.; Wang, H.C. The regulation of shrimp metabolism by the white spot syndrome virus (WSSV). Rev. Aquac. 2022, 14, 1150–1169. [Google Scholar] [CrossRef]
- Dashtiannasab, A. White spot syndrome virus. In Emerging and Reemerging Viral Pathogens; Elsevier: Amsterdam, The Netherlands, 2020; pp. 717–728. [Google Scholar]
- Escobedo-Bonilla, C.M.; Alday-Sanz, V.; Wille, M.; Sorgeloos, P.; Pensaert, M.; Nauwynck, H. A review on the morphology, molecular characterization, morphogenesis and pathogenesis of white spot syndrome virus. J. Fish Dis. 2008, 31, 1–18. [Google Scholar] [CrossRef]
- Feng, S.; Wang, C.; Hu, S.; Wu, Q.; Li, A. Recent progress in the development of white spot syndrome virus vaccines for protecting shrimp against viral infection. Arch. Virol. 2017, 162, 2923–2936. [Google Scholar] [CrossRef]
- Witteveldt, J.; Vlak, J.M.; van Hulten, M.C. Protection of Penaeus monodon against white spot syndrome virus using a WSSV subunit vaccine. Fish Shellfish Immunol. 2004, 16, 571–579. [Google Scholar] [CrossRef]
- Gao, G.; Lin, R.; Tao, M.; Aweya, J.J.; Yao, D.; Ma, H.; Li, S.; Zhang, Y.; Wang, F. Molecular characterization of a novel white spot syndrome virus response protein (dubbed LvWRP) from Litopenaeus vannamei. Dev. Comp. Immunol. 2019, 98, 99–107. [Google Scholar] [CrossRef]
- Phanse, Y.; Puttamreddy, S.; Loy, D.; Ramirez, J.V.; Ross, K.A.; Alvarez-Castro, I.; Mogler, M.; Broderick, S.; Rajan, K.; Narasimhan, B. RNA Nanovaccine Protects against White Spot Syndrome Virus in Shrimp. Vaccines 2022, 10, 1428. [Google Scholar] [CrossRef]
- Abreu, A.P.; Fernandes, B.; Vicente, A.A.; Teixeira, J.; Dragone, G. Mixotrophic cultivation of Chlorella vulgaris using industrial dairy waste as organic carbon source. Bioresour. Technol. 2012, 118, 61–66. [Google Scholar] [CrossRef] [PubMed]
- Yang, B.; Liu, J.; Jiang, Y.; Chen, F. Chlorella species as hosts for genetic engineering and expression of heterologous proteins: Progress, challenge and perspective. Biotechnol. J. 2016, 11, 1244–1261. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Li, X.; Yao, W.; Wang, Y.; Zhang, X.; Leng, X. An Evaluation of Replacing Fishmeal with Chlorella Sorokiniana in the Diet of Pacific White Shrimp (Litopenaeus Vannamei): Growth, Body Color, and Flesh Quality. Aquac. Nutr. 2022, 2022, 8617265. [Google Scholar] [CrossRef] [PubMed]
- Dehghani, J.; Movafeghi, A.; Mathieu-Rivet, E.; Mati-Baouche, N.; Calbo, S.; Lerouge, P.; Bardor, M. Microalgae as an efficient vehicle for the production and targeted delivery of therapeutic glycoproteins against SARS-CoV-2 variants. Mar. Drugs 2022, 20, 657. [Google Scholar] [CrossRef]
- Kim, M.-J.; Kim, S.-H.; Kim, Y.-R.; Choi, T.-J. Enhancement of Chlorella transformation efficacy by insert fragmentation. Algal Res. 2023, 72, 103146. [Google Scholar] [CrossRef]
- Jeeva, S.; Kim, N.I.; Jang, I.K.; Choi, T.J. Development of a multiplex PCR system for the simultaneous detection of white spot syndrome virus and hepatopancreatic parvovirus infection. Aquac. Res. 2014, 45, 1073–1083. [Google Scholar] [CrossRef]
- Durand, S.; Redman, R.; Mohney, L.; Tang-Nelson, K.; Bonami, J.; Lightner, D. Qualitative and quantitative studies on the relative virus load of tails and heads of shrimp acutely infected with WSSV. Aquaculture 2003, 216, 9–18. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Raguraman, V.; Ravindran, N.; Selvaraju, K.; Kasivelu, G. Seaweed polysaccharides as potential therapeutic agents against white spot syndrome virus (WSSV): A mini review. Aquac. Int. 2020, 28, 2333–2343. [Google Scholar] [CrossRef]
- Dekham, K.; Jitrakorn, S.; Charoonnart, P.; Isarangkul, D.; Chaturongakul, S.; Saksmerprome, V. Probiotics expressing double-stranded RNA targeting VP28 efficiently protect shrimps from WSSV infection. Aquac. Rep. 2022, 23, 101067. [Google Scholar] [CrossRef]
- Van Hulten, M.C.; Westenberg, M.; Goodall, S.D.; Vlak, J.M. Identification of two major virion protein genes of white spot syndrome virus of shrimp. Virology 2000, 266, 227–236. [Google Scholar] [CrossRef]
- Feng, S.; Feng, W.; Zhao, L.; Gu, H.; Li, Q.; Shi, K.; Guo, S.; Zhang, N. Preparation of transgenic Dunaliella salina for immunization against white spot syndrome virus in crayfish. Arch. Virol. 2014, 159, 519–525. [Google Scholar] [CrossRef]
- Caipang, C.M.A.; Verjan, N.; Ooi, E.L.; Kondo, H.; Hirono, I.; Aoki, T.; Kiyono, H.; Yuki, Y. Enhanced survival of shrimp, Penaeus (Marsupenaeus) japonicus from white spot syndrome disease after oral administration of recombinant VP28 expressed in Brevibacillus brevis. Fish Shellfish Immunol. 2008, 25, 315–320. [Google Scholar] [CrossRef]
- Fu, L.-L.; Shuai, J.-B.; Xu, Z.-R.; Li, J.-R.; Li, W.-F. Immune responses of Fenneropenaeus chinensis against white spot syndrome virus after oral delivery of VP28 using Bacillus subtilis as vehicles. Fish Shellfish Immunol. 2010, 28, 49–55. [Google Scholar] [CrossRef]
- Yang, K.H.; Lee, M.G. Effects of endotoxin derived from Escherichia coli lipopolysaccharide on the pharmacokinetics of drugs. Arch. Pharmacal Res. 2008, 31, 1073–1086. [Google Scholar] [CrossRef]
- Mohammadi, G.; Adorian, T.J.; Rafiee, G. Beneficial effects of Bacillus subtilis on water quality, growth, immune responses, endotoxemia and protection against lipopolysaccharide-induced damages in Oreochromis niloticus under biofloc technology system. Aquac. Nutr. 2020, 26, 1476–1492. [Google Scholar] [CrossRef]
- Widyaningrum, D.; Prianto, A.D. Chlorella as a source of functional food ingredients: Short review. In Proceedings of the IOP Conference Series: Earth and Environmental Science; IOP Publishing: Bristol, UK, 2021; p. 012148. [Google Scholar]
- Dehghani, J.; Adibkia, K.; Movafeghi, A.; Maleki-Kakelar, H.; Saeedi, N.; Omidi, Y. Towards a new avenue for producing therapeutic proteins: Microalgae as a tempting green biofactory. Biotechnol. Adv. 2020, 40, 107499. [Google Scholar] [CrossRef]
- Dona, A.; Arvanitoyannis, I.S. Health risks of genetically modified foods. Crit. Rev. Food Sci. Nutr. 2009, 49, 164–175. [Google Scholar] [CrossRef]
- Hossain, A.; Habibullah-Al-Mamun, M.; Nagano, I.; Masunaga, S.; Kitazawa, D.; Matsuda, H. Antibiotics, antibiotic-resistant bacteria, and resistance genes in aquaculture: Risks, current concern, and future thinking. Environ. Sci. Pollut. Res. 2022, 29, 11054–11075. [Google Scholar] [CrossRef]
- Xie, X.; Xu, L.; Yang, F. Proteomic analysis of the major envelope and nucleocapsid proteins of white spot syndrome virus. J. Virol. 2006, 80, 10615–10623. [Google Scholar] [CrossRef]
- Chu, F.; Cheng, J.; Li, K.; Wang, Y.; Li, X.; Yang, W. Enhanced lipid accumulation through a regulated metabolic pathway of phosphorus luxury uptake in the microalga Chlorella vulgaris under nitrogen starvation and phosphorus repletion. ACS Sustain. Chem. Eng. 2020, 8, 8137–8147. [Google Scholar] [CrossRef]
- Choi, J.; Shin, J.-H.; An, H.J.; Oh, M.J.; Kim, S.-R. Analysis of secretome and N-glycosylation of Chlorella species. Algal Res. 2021, 59, 102466. [Google Scholar] [CrossRef]
- Li, X.; Liu, Q.-H.; Hou, L.; Huang, J. Effect of VP28 DNA vaccine on white spot syndrome virus in Litopenaeus vannamei. Aquac. Int. 2010, 18, 1035–1044. [Google Scholar] [CrossRef]
- Jha, R.K.; Xu, Z.R.; Shen, J.; Bai, S.J.; Sun, J.Y.; Li, W.F. The efficacy of recombinant vaccines against white spot syndrome virus in Procambarus clarkii. Immunol. Lett. 2006, 105, 68–76. [Google Scholar] [CrossRef]
- Zhai, Y.-F.; Shi, D.-J.; He, P.-M.; Cai, C.-E.; Yin, R.; Jia, R. Effect of trans-vp28 gene Synechocystis sp. PCC6803 on growth and immunity of Litopenaeus vannamei and defense against white spot syndrome virus (WSSV). Aquaculture 2019, 512, 734306. [Google Scholar] [CrossRef]
- McLean, E.; Barrows, F.; Craig, S.; Alfrey, K.; Tran, L. Complete replacement of fishmeal by soybean and poultry meals in Pacific whiteleg shrimp feeds: Growth and tolerance to EMS/AHPND and WSSV challenge. Aquaculture 2020, 527, 735383. [Google Scholar] [CrossRef]
- Lanh, P.T.; Nguyen, H.M.; Duong, B.T.T.; Hoa, N.T.; Tam, L.T.; Thu, H.T.; Van Nha, V.; Hong, D.D.; Mouradov, A.; Koyande, A.K. Generation of microalga Chlamydomonas reinhardtii expressing VP28 protein as oral vaccine candidate for shrimps against White Spot Syndrome Virus (WSSV) infection. Aquaculture 2021, 540, 736737. [Google Scholar] [CrossRef]
- Kumar, S.R.; Ahamed, V.I.; Sarathi, M.; Basha, A.N.; Hameed, A.S. Immunological responses of Penaeus monodon to DNA vaccine and its efficacy to protect shrimp against white spot syndrome virus (WSSV). Fish Shellfish Immunol. 2008, 24, 467–478. [Google Scholar] [CrossRef]
- Fajardo, C.; Martinez-Rodriguez, G.; Costas, B.; Mancera, J.M.; Fernandez-Boo, S.; Rodulfo, H.; De Donato, M. Shrimp immune response: A transcriptomic perspective. Rev. Aquac. 2022, 14, 1136–1149. [Google Scholar] [CrossRef]
- Janeway, C.A., Jr.; Medzhitov, R. Innate immune recognition. Annu. Rev. Immunol. 2002, 20, 197–216. [Google Scholar] [CrossRef]
- Wang, P.-H.; Huang, T.; Zhang, X.; He, J.-G. Antiviral defense in shrimp: From innate immunity to viral infection. Antivir. Res. 2014, 108, 129–141. [Google Scholar] [CrossRef]
- Amparyup, P.; Charoensapsri, W.; Tassanakajon, A. Prophenoloxidase system and its role in shrimp immune responses against major pathogens. Fish Shellfish Immunol. 2013, 34, 990–1001. [Google Scholar] [CrossRef]
- Liu, J.; Yu, Y.; Li, F.; Zhang, X.; Xiang, J. A new anti-lipopolysaccharide factor (ALF) gene with its SNP polymorphisms related to WSSV-resistance of Litopenaeus vannamei. Fish Shellfish Immunol. 2014, 39, 24–33. [Google Scholar] [CrossRef]
- De la Vega, E.; O’Leary, N.A.; Shockey, J.E.; Robalino, J.; Payne, C.; Browdy, C.L.; Warr, G.W.; Gross, P.S. Anti-lipopolysaccharide factor in Litopenaeus vannamei (LvALF): A broad spectrum antimicrobial peptide essential for shrimp immunity against bacterial and fungal infection. Mol. Immunol. 2008, 45, 1916–1925. [Google Scholar] [CrossRef]
- Viana, J.T.; dos Santos Rocha, R.; Maggioni, R. Structural and functional diversity of lectins associated with immunity in the marine shrimp Litopenaeus vannamei. Fish Shellfish Immunol. 2022, 129, 152–160. [Google Scholar] [CrossRef]
- Li, M.; Li, C.; Ma, C.; Li, H.; Zuo, H.; Weng, S.; Chen, X.; Zeng, D.; He, J.; Xu, X. Identification of a C-type lectin with antiviral and antibacterial activity from pacific white shrimp Litopenaeus vannamei. Dev. Comp. Immunol. 2014, 46, 231–240. [Google Scholar] [CrossRef]
- Wang, X.-W.; Wang, J.-X. Diversity and multiple functions of lectins in shrimp immunity. Dev. Comp. Immunol. 2013, 39, 27–38. [Google Scholar] [CrossRef]
- Wang, C.; Wei, M.; Wu, G.; He, L.; Zhu, J.; Juventus Aweya, J.; Chen, X.; Zhao, Y.; Zhang, Y.; Yao, D. Proteomics analysis reveals a critical role for the WSSV immediate-early protein IE1 in modulating the host prophenoloxidase system. Virulence 2022, 13, 936–948. [Google Scholar] [CrossRef]





| Target | Primer Name | Sequences | PCR Product |
|---|---|---|---|
| 1-NR | 1-NR F | 5′- ATGGACAAGACAGGGTTCGG -3′ | 2556 bp |
| 1-NR R | 5′- CAACGAGCGCCTCCATTTAC -3′ | ||
| 2-NR | 2-NR F | 5′- CACGAGGAGCATCGTGGAAA -3 | 2065 bp |
| 2-NR R | 5′- AATACAGGCGGAGCCCAAAC -3′ | ||
| VP28 | VP28 F | 5′- TCGCTACCACAATACGGTG -3′ | 483 bp |
| VP28 R | 5′- TTGCCACCGGCTGTTG -3′ | ||
| WSSV | WSSV F | 5′- CTTTCACTCTTTCGGTCGTGTC -3′ | 604 bp |
| WSSV R | 5′- TACTCGGTCTCAGTGCCAGA -3′ | ||
| Nested- WSSV | Nested F | 5′- CCCACACAGACAATATCGAGACAA -3′ | 258 bp |
| Nested R | 5′- CTTGATGTGTTGTTCCACACCTTG -3′ | ||
| qWSSV | qWSSV F | 5′- ATCCTCGCCATCACTGCTGT -3′ | 560 bp |
| qWSSV R | 5′- GAGTAGGTGACGTGCACGTA -3′ | ||
| qVP28 | qVP28 F | 5′- GTGACCAAGACCATCGAAAC –3′ | 110 bp |
| qVP28 R | 5′- TCAGTCATCTTGAAGTAGCC -3′ | ||
| ALF | ALF F | 5′- CCTCATCCCTTCGCTAGTCCA -3′ | 148 bp |
| ALF R | 5′- CCATCCAGGACACCACATCC -3′ | ||
| Lectin | Lectin F | 5′- TTACTCTCGGTGTCGTTGGGAC -3′ | 148 bp |
| Lectin R | 5′- ATCTGATACCAGGTTCCCTCAGTC -3′ | ||
| proPO | proPO F | 5′- TATGAAGACGAACGGGGCC -3′ | 133 bp |
| proPO R | 5′- CAGACGGACGGAATGGAGTT -3′ | ||
| β-actin | β-actin F | 5′- ATGTTCGAGACCTTCAACACCC -3′ | 135 bp |
| β-actin R | 5′- CTCGTAGATGGGCACGGT -3′ |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, M.-J.; Kim, S.-H.; Kim, J.-O.; Lee, T.-K.; Jang, I.-K.; Choi, T.-J. Efficacy of White Spot Syndrome Virus Protein VP28-Expressing Chlorella vulgaris as an Oral Vaccine for Shrimp. Viruses 2023, 15, 2010. https://doi.org/10.3390/v15102010
Kim M-J, Kim S-H, Kim J-O, Lee T-K, Jang I-K, Choi T-J. Efficacy of White Spot Syndrome Virus Protein VP28-Expressing Chlorella vulgaris as an Oral Vaccine for Shrimp. Viruses. 2023; 15(10):2010. https://doi.org/10.3390/v15102010
Chicago/Turabian StyleKim, Min-Jeong, Su-Hyun Kim, Jong-Oh Kim, Taek-Kyun Lee, In-Kwon Jang, and Tae-Jin Choi. 2023. "Efficacy of White Spot Syndrome Virus Protein VP28-Expressing Chlorella vulgaris as an Oral Vaccine for Shrimp" Viruses 15, no. 10: 2010. https://doi.org/10.3390/v15102010
APA StyleKim, M. -J., Kim, S. -H., Kim, J. -O., Lee, T. -K., Jang, I. -K., & Choi, T. -J. (2023). Efficacy of White Spot Syndrome Virus Protein VP28-Expressing Chlorella vulgaris as an Oral Vaccine for Shrimp. Viruses, 15(10), 2010. https://doi.org/10.3390/v15102010