The Way of Water: Unravelling White Spot Syndrome Virus (WSSV) Transmission Dynamics in Litopenaeus vannamei Shrimp
Abstract
:1. Introduction
2. Materials and Methods
2.1. Experimental Animals and Rearing Conditions
2.2. Experimental Conditions
2.3. WSSV Stock and Peroral Inoculation
2.4. In Vivo WSSV Challenges
2.4.1. Characterization of WSD Progression
2.4.2. WSSV Infection in Relation to Population Density
2.4.3. The Effect of Isolation on WSSV Epidemic Dynamics
2.4.4. Role of Molted Cuticles, Feces, and Rearing Water in WSSV Transmission Dynamics
2.5. Confirmation of Viral Infection and Shedding
2.6. Statistical Analysis
3. Results
3.1. In Vivo Titrations of WSSV-Positive Tissue Inocula
3.2. In Vivo WSSV Challenges
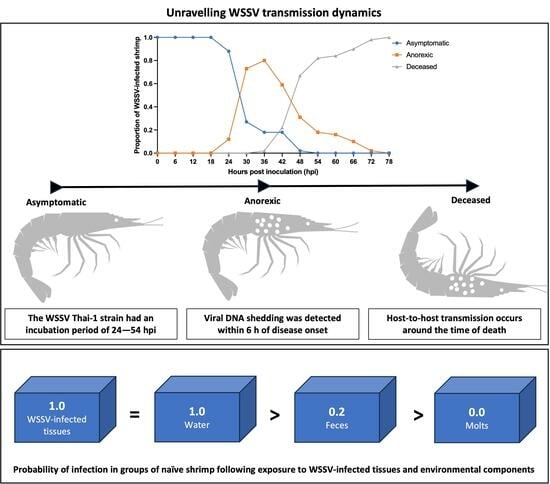
3.2.1. Characterization of WSD Progression
3.2.2. WSSV Infection in Relation to Population Density
3.2.3. The Effect of Isolation on WSSV Epidemic Dynamics
3.2.4. Role of Molted Cuticles, Feces, and Rearing Water in WSSV Transmission Dynamics
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chamberlain, G.; Lightner, D.; Towner, R.; van Wyk, P.; Villarreal, M.; Akazawa, N.; Alvial, A. Case Study of the Outbreak of White Spot Syndrome Virus at Shrimp Farms in Mozambique and Madagascar: Impacts and Management Recommendations. Responsible Aquac. Organ. 2013. [Google Scholar]
- Brummett, R.E.; Alvial, A.; Kibenge, F.; Forster, J.; Burgos, J.M.; Ibarra, R.; St-Hilaire, S.; Chamberlain, G.C.; Lightner, D.V.; Van Khoa, L.; et al. Reducing Disease Risk In Aquaculture; The World Bank: Washington, DC, USA, 2014. [Google Scholar]
- Anderson, J.L.; Asche, F.; Garlock, T. Economics of Aquaculture Policy and Regulation. Annu. Rev. Resour. Econ. 2019, 11, 101–123. [Google Scholar] [CrossRef]
- Pradeep, B.; Rai, P.; Mohan, S.A.; Shekhar, M.S.; Karunasagar, I. Biology, Host Range, Pathogenesis and Diagnosis of White Spot Syndrome Virus. Indian J. Virol. 2012, 23, 161–174. [Google Scholar] [CrossRef]
- Chou, H.; Huang, C.; Wang, C.; Chiang, H.; Lo, C. Pathogenicity of a Baculovirus Infection Causing White Spot Syndrome in Cultured Penaeid Shrimp in Taiwan. Dis. Aquat. Org. 1995, 23, 165–173. [Google Scholar] [CrossRef]
- OIE. Manual of Diagnostic Tests for Aquatic Animals; Office International des Épizooties: Paris, France, 2003; Volume 33, ISBN 9290445637. [Google Scholar]
- Pradeep, B.; Shekar, M.; Karunasagar, I.; Karunasagar, I. Characterization of Variable Genomic Regions of Indian White Spot Syndrome Virus. Virology 2008, 376, 24–30. [Google Scholar] [CrossRef] [PubMed]
- Joseph, T.C.; James, R.; Rajan, L.A.; Surendran, P.K.; Lalitha, K.V. White Spot Syndrome Virus Infection: Threat to Crustacean Biodiversity in Vembanad Lake, India. Biotechnol. Rep. 2015, 7, 51–54. [Google Scholar] [CrossRef]
- Dey, B.K.; Dugassa, G.H.; Hinzano, S.M.; Bossier, P. Causative Agent, Diagnosis and Management of White Spot Disease in Shrimp: A Review. Rev. Aquac. 2020, 12, 822–865. [Google Scholar] [CrossRef]
- Verbruggen, B.; Bickley, L.K.; van Aerle, R.; Bateman, K.S.; Stentiford, G.D.; Santos, E.M.; Tyler, C.R. Molecular Mechanisms of White Spot Syndrome Virus Infection and Perspectives on Treatments. Viruses 2016, 8, 23. [Google Scholar] [CrossRef] [PubMed]
- Desrina; Prayitno, S.B.; Verdegem, M.C.J.; Verreth, J.A.J.; Vlak, J.M. White Spot Syndrome Virus Host Range and Impact on Transmission. Rev. Aquac. 2022, 14, 1843–1860. [Google Scholar] [CrossRef]
- Lo, C.; Ho, C.; Chen, C.; Liu, K.; Chiu, Y.; Yeh, P.; Peng, S.; Hsu, H.; Liu, H.; Chang, C.; et al. Detection and Tissue Tropism of White Spot Syndrome Baculovirus (WSBV) in Captured Brooders of Penaeus Monodon with a Special Emphasis on Reproductive Organs. Dis. Aquat. Org. 1997, 30, 53–72. [Google Scholar] [CrossRef]
- Chang, P.S.; Lo, C.F.; Wang, Y.C.; Kou, G.H. Identification of White Spot Syndrome Associated Baculovirus (WSBV) Target Organs in the Shrimp Penaeus Monodon by in Situ Hybridization. Dis. Aquat. Org. 1996, 27, 131–139. [Google Scholar] [CrossRef]
- Zhang, J.S.; Dong, S.L.; Tian, X.L.; Dong, Y.W.; Liu, X.Y.; Yan, D.C. Studies on the Rotifer (Brachionus Urceus Linnaeus, 1758) as a Vector in White Spot Syndrome Virus (WSSV) Transmission. Aquaculture 2006, 261, 1181–1185. [Google Scholar] [CrossRef]
- Tuyen, N.X.; Verreth, J.; Vlak, J.M.; de Jong, M.C.M. Horizontal Transmission Dynamics of White Spot Syndrome Virus by Cohabitation Trials in Juvenile Penaeus Monodon and P. Vannamei. Prev. Vet. Med. 2014, 117, 286–294. [Google Scholar] [CrossRef] [PubMed]
- Domínguez-Borbor, C.; Betancourt, I.; Panchana, F.; Sonnenholzner, S.; Bayot, B. An Effective White Spot Syndrome Virus Challenge Test for Cultured Shrimp Using Different Biomass of the Infected Papilla. MethodsX 2019, 6, 1617. [Google Scholar] [CrossRef] [PubMed]
- Soto, M.A.; Lotz, J.M. Epidemiological Parameters of White Spot Syndrome Virus Infections in Litopenaeus Vannamei and L. Setiferus. J. Invertebr. Pathol. 2001, 78, 9–15. [Google Scholar] [CrossRef] [PubMed]
- Lotz, J.M.; Soto, M.A. Model of White Spot Syndrome Virus (WSSV) Epidemics in Litopenaeus Vannamei. Dis. Aquat. Org. 2002, 50, 199–209. [Google Scholar] [CrossRef]
- Jewell, N.P. Natural History of Diseases: Statistical Designs and Issues. Clin. Pharmacol. Ther. 2016, 100, 353. [Google Scholar] [CrossRef]
- De La Paz, M.P.; Villaverde-Hueso, A.; Alonso, V.; János, S.; Zurriaga, Ó.; Pollán, M.; Abaitua-Borda, I. Rare Diseases Epidemiology Research. Adv. Exp. Med. Biol. 2010, 686, 17–39. [Google Scholar] [CrossRef]
- Jiravanichpaisal, P.; Söderhäll, K.; Söderhäll, I. Effect of Water Temperature on the Immune Response and Infectivity Pattern of White Spot Syndrome Virus (WSSV) in Freshwater Crayfish. Fish Shellfish Immunol. 2004, 17, 265–275. [Google Scholar] [CrossRef]
- Thuong, K.; Tuan, V.; Li, W.; Sorgeloos, P.; Bossier, P.; Nauwynck, H. Per Os Infectivity of White Spot Syndrome Virus (WSSV) in White-Legged Shrimp (Litopenaeus Vannamei) and Role of Peritrophic Membrane. Vet. Res. 2016, 47, 39. [Google Scholar] [CrossRef]
- Esparza-Leal, H.M.; Escobedo-Bonilla, C.M.; Casillas-Hernández, R.; Álvarez-Ruíz, P.; Portillo-Clark, G.; Valerio-García, R.C.; Hernández-López, J.; Méndez-Lozano, J.; Vibanco-Pérez, N.; Magallón-Barajas, F.J. Detection of White Spot Syndrome Virus in Filtered Shrimp-Farm Water Fractions and Experimental Evaluation of Its Infectivity in Penaeus (Litopenaeus) Vannamei. Aquaculture 2009, 292, 16–22. [Google Scholar] [CrossRef]
- Satheesh Kumar, S.; Ananda Bharathi, R.; Rajan, J.J.S.; Alavandi, S.V.; Poornima, M.; Balasubramanian, C.P.; Ponniah, A.G. Viability of White Spot Syndrome Virus (WSSV) in Sediment during Sun-Drying (Drainable Pond) and under Non-Drainable Pond Conditions Indicated by Infectivity to Shrimp. Aquaculture 2013, 402–403, 119–126. [Google Scholar] [CrossRef]
- Pérez, F.; Volckaert, F.A.M.; Calderón, J. Pathogenicity of White Spot Syndrome Virus on Postlarvae and Juveniles of Penaeus (Litopenaeus) Vannamei. Aquaculture 2005, 250, 586–591. [Google Scholar] [CrossRef]
- Waikhom, G.; John, K.R.; George, M.R.; Jeyaseelan, M.J.P. Differential Host Passaging Alters Pathogenicity and Induces Genomic Variation in White Spot Syndrome Virus. Aquaculture 2006, 261, 54–63. [Google Scholar] [CrossRef]
- John, K.R.; George, M.R.; Iyappan, T.; Thangarani, A.J.; Jeyaseelan, M.J.P. Indian Isolates of White Spot Syndrome Virus Exhibit Variations in Their Pathogenicity and Genomic Tandem Repeats. J. Fish Dis. 2010, 33, 749–758. [Google Scholar] [CrossRef] [PubMed]
- Sahul Hameed, A.S.; Anilkumar, M.; Raj, M.L.S.; Jayaraman, K. Studies on the Pathogenicity of Systemic Ectodermal and Mesodermal Baculovirus and Its Detection in Shrimp by Immunological Methods. Aquaculture 1998, 160, 31–45. [Google Scholar] [CrossRef]
- Bateman, K.S.; Tew, I.; French, C.; Hicks, R.J.; Martin, P.; Munro, J.; Stentiford, G.D. Susceptibility to Infection and Pathogenicity of White Spot Disease (WSD) in Non-Model Crustacean Host Taxa from Temperate Regions. J. Invertebr. Pathol. 2012, 110, 340–351. [Google Scholar] [CrossRef] [PubMed]
- Pradeep, B.; Karunasagar, I.; Karunasagar, I. Fitness and Virulence of Different Strains of White Spot Syndrome Virus. J. Fish Dis. 2009, 32, 801–805. [Google Scholar] [CrossRef] [PubMed]
- Marks, H.; Van Duijse, J.J.A.; Zuidema, D.; Van Hulten, M.C.W.; Vlak, J.M. Fitness and Virulence of an Ancestral White Spot Syndrome Virus Isolate from Shrimp. Virus Res. 2005, 110, 9–20. [Google Scholar] [CrossRef]
- Li, F.; Gao, M.; Xu, L.; Yang, F. Comparative Genomic Analysis of Three White Spot Syndrome Virus Isolates of Different Virulence. Virus Genes 2017, 53, 249–258. [Google Scholar] [CrossRef]
- Gao, M.; Li, F.; Xu, L.; Zhu, X. White Spot Syndrome Virus Strains of Different Virulence Induce Distinct Immune Response in Cherax Quadricarinatus. Fish Shellfish Immunol. 2014, 39, 17–23. [Google Scholar] [CrossRef] [PubMed]
- Ramos-Paredes, J.; Grijalva-Chon, J.M.; Ibarra-Gámez, J.C. Virulence and Genotypes of White Spot Syndrome Virus Infecting Pacific White Shrimp Litopenaeus Vannamei in North-Western Mexico. J. Fish Dis. 2017, 40, 425–435. [Google Scholar] [CrossRef]
- Corteel, M.; Dantas-Lima, J.J.; Tuan, V.V.; Van Thuong, K.; Wille, M.; Alday-Sanz, V.; Pensaert, M.B.; Sorgeloos, P.; Nauwynck, H.J. Susceptibility of Juvenile Macrobrachium Rosenbergii to Different Doses of High and Low Virulence Strains of White Spot Syndrome Virus (WSSV). Dis. Aquat. Org. 2012, 100, 211–218. [Google Scholar] [CrossRef] [PubMed]
- Rahman, M.M.; Corteel, M.; Escobedo-Bonilla, C.M.; Wille, M.; Alday-Sanz, V.; Pensaert, M.B.; Sorgeloos, P.; Nauwynck, H.J. Virulence of White Spot Syndrome Virus (WSSV) Isolates May Be Correlated with the Degree of Replication in Gills of Penaeus Vannamei Juveniles. Dis. Aquat. Org. 2008, 79, 191–198. [Google Scholar] [CrossRef] [PubMed]
- Straif-Bourgeois, S.; Ratard, R.; Kretzschmar, M. Infectious Disease Epidemiology. Handb. Epidemiol. 2014, 2041–2119. [Google Scholar] [CrossRef]
- Giesecke, J. Modern Infectious Disease Epidemiology: Third Edition. Mod. Infect. Dis. Epidemiol. Third Ed. 2017, 1–233. [Google Scholar] [CrossRef]
- Brachman, P.S. Epidemiology. Med. Microbiol. 1996, 378. [Google Scholar]
- Zhang, R.; Li, Y.; Zhang, A.L.; Wang, Y.; Molina, M.J. Identifying Airborne Transmission as the Dominant Route for the Spread of COVID-19. Proc. Natl. Acad. Sci. USA 2020, 117, 14857–14863. [Google Scholar] [CrossRef]
- Escobedo-Bonilla, C.M.; Wille, M.; Alday Sanz, V.; Sorgeloos, P.; Pensaert, M.B.; Nauwynck, H.J. Pathogenesis of a Thai Strain of White Spot Syndrome Virus (WSSV) in Juvenile, Specific Pathogen-Free Litopenaeus Vannamei. Dis. Aquat. Org. 2007, 74, 85–94. [Google Scholar] [CrossRef]
- Escobedo-Bonilla, C.M.; Audoorn, L.; Wille, M.; Alday-Sanz, V.; Sorgeloos, P.; Pensaert, M.B.; Nauwynck, H.J. Standardized White Spot Syndrome Virus (WSSV) Inoculation Procedures for Intramuscular or Oral Routes. Dis. Aquat. Org. 2006, 68, 181–188. [Google Scholar] [CrossRef]
- De Gryse, G.M.A.; Khuong, T.V.; Descamps, B.; Van Den Broeck, W.; Vanhove, C.; Cornillie, P.; Sorgeloos, P.; Bossier, P.; Nauwynck, H.J. The Shrimp Nephrocomplex Serves as a Major Portal of Pathogen Entry and Is Involved in the Molting Process. Proc. Natl. Acad. Sci. USA 2020, 117, 28374. [Google Scholar] [CrossRef]
- Corteel, M.; Nauwynck, H.J.; Sorgeloos, P.; Alday-Sanz, V. White Spot Syndrome Virus Infection in P. Vannamei and M. Rosenbergii: Experimental Studies on Susceptibility to Infection and Disease; Faculty of Veterinary Medicine, Ghent University: Merelbeke, Belgium, 2013; ISBN 9789058643308. [Google Scholar]
- Richardson, A.; Dantas-Lima, J.; Lefranc, M.; Walraven, M. Effect of a Black Soldier Fly Ingredient on the Growth Performance and Disease Resistance of Juvenile Pacific White Shrimp (Litopenaeus Vannamei). Animals 2021, 11, 1450. [Google Scholar] [CrossRef] [PubMed]
- Escobedo-Bonilla, C.M.; Wille, M.; Alday Sanz, V.; Sorgeloos, P.; Pensaert, M.B.; Nauwynck, H.J. In Vivo Titration of White Spot Syndrome Virus (WSSV) in Specific Pathogen-Free Litopenaeus Vannamei by Intramuscular and Oral Routes. Dis. Aquat. Org. 2005, 66, 163–170. [Google Scholar] [CrossRef]
- Jiravanichpaisal, P.; Bangyeekhun, E.; Söderhäll, K.; Söderhäll, I. Experimental Infection of White Spot Syndrome Virus in Freshwater Crayfish Pacifastacus Leniusculus. Dis. Aquat. Org. 2001, 47, 151–157. [Google Scholar] [CrossRef] [PubMed]
- Pahuta, M.A.; Werier, J.; Wai, E.K.; Patchell, R.A.; Coyle, D. A Technique for Approximating Transition Rates from Published Survival Analyses. Cost Eff. Resour. Alloc. 2019, 17, 12. [Google Scholar] [CrossRef]
- Djurhuus, A.; Port, J.; Closek, C.J.; Yamahara, K.M.; Romero-Maraccini, O.; Walz, K.R.; Goldsmith, D.B.; Michisaki, R.; Breitbart, M.; Boehm, A.B.; et al. Evaluation of Filtration and DNA Extraction Methods for Environmental DNA Biodiversity Assessments across Multiple Trophic Levels. Front. Mar. Sci. 2017, 4, 314. [Google Scholar] [CrossRef]
- Li, W.; Desmarets, L.M.B.; De Gryse, G.M.A.; Theuns, S.; Van Tuan, V.; Van Thuong, K.; Bossier, P.; Nauwynck, H.J. Virus Replication Cycle of White Spot Syndrome Virus in Secondary Cell Cultures from the Lymphoid Organ of Litopenaeus Vannamei. J. Gen. Virol. 2015, 96, 2844–2854. [Google Scholar] [CrossRef]
- Corteel, M.; Dantas-Lima, J.J.; Wille, M.; Alday-Sanz, V.; Pensaert, M.B.; Sorgeloos, P.; Nauwynck, H.J. Molt Stage and Cuticle Damage Influence White Spot Syndrome Virus Immersion Infection in Penaeid Shrimp. Vet. Microbiol. 2009, 137, 209–216. [Google Scholar] [CrossRef]
- Comer, J.E.; Brasel, T.; Massey, S.; Beasley, D.W.; Cirimotich, C.M.; Sanford, D.C.; Chou, Y.L.; Niemuth, N.A.; Novak, J.; Sabourin, C.L.; et al. Natural History of Marburg Virus Infection to Support Medical Countermeasure Development. Viruses 2022, 14, 2291. [Google Scholar] [CrossRef]
- Kim, M.-J.; Kim, J.-O.; Jang, G.-I.; Kwon, M.-G.; Kim, K.-I. Evaluation of the Horizontal Transmission of White Spot Syndrome Virus for Whiteleg Shrimp (Litopenaeus Vannamei) Based on the Disease Severity Grade and Viral Shedding Rate. Animals 2023, 13, 1676. [Google Scholar] [CrossRef]
- Quang, N.D.; Hoa, P.T.P.; Da, T.T.; Anh, P.H. Persistence of White Spot Syndrome Virus in Shrimp Ponds and Surrounding Areas after an Outbreak. Environ. Monit. Assess. 2009, 156, 69–72. [Google Scholar] [CrossRef]
- Tendencia, E.A.; Bosma, R.H.; Verreth, J.A.J. White Spot Syndrome Virus (WSSV) Risk Factors Associated with Shrimp Farming Practices in Polyculture and Monoculture Farms in the Philippines. Aquaculture 2011, 311, 87–93. [Google Scholar] [CrossRef]
- Wu, J.L.; Namikoshi, A.; Nishizawa, T.; Mushiake, K.; Teruya, K.; Muroga, K. Effects of Shrimp Density on Transmission of Penaeid Acute Viremia in Penaeus Japonicus by Cannibalism and the Waterborne Route. Dis. Aquat. Org. 2001, 47, 129–135. [Google Scholar] [CrossRef] [PubMed]
- Kautsky, N.; Rönnbäck, P.; Tedengren, M.; Troell, M. Ecosystem Perspectives on Management of Disease in Shrimp Pond Farming. Aquaculture 2000, 191, 145–161. [Google Scholar] [CrossRef]
- Ruiz-Velazco, J.M.J.; Hernández-Llamas, A.; Gomez-Muñoz, V.M.; Magallon, F.J. Dynamics of Intensive Production of Shrimp Litopenaeus Vannamei Affected by White Spot Disease. Aquaculture 2010, 300, 113–119. [Google Scholar] [CrossRef]
- Apún-Molina, J.P.; Robles-Romo, A.; Alvarez-Ruiz, P.; Santamaria-Miranda, A.; Arjona, O.; Racotta, I.S. Influence of Stocking Density and Exposure to White Spot Syndrome Virus in Biological Performance, Metabolic, Immune, and Bioenergetics Response of Whiteleg Shrimp Litopenaeus Vannamei. Aquaculture 2017, 479, 528–537. [Google Scholar] [CrossRef]
- Ojoma, A.Q.; David, O.; Chukwuma, A.C.; Ugochukwu, N.D.; Ezike, M.G.C. Transmission Dynamics of Ebola Virus Disease with Vaccine, Condom Use, Quarantine, Isolation and Treatment Drug. Afr. J. Infect. Dis. 2020, 15, 10–23. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.; Lao, A. Transmission Dynamics and Control Strategies Assessment of Avian Influenza A (H5N6) in the Philippines. Infect. Dis. Model. 2018, 3, 35–59. [Google Scholar] [CrossRef] [PubMed]
- Sheng, Y.; Cui, J.A.; Guo, S. The Modeling and Analysis of the COVID-19 Pandemic with Vaccination and Isolation: A Case Study of Italy. Math. Biosci. Eng. 2023, 20, 5966–5992. [Google Scholar] [CrossRef]
- Behera, B.K.; Prasad, R. Shyambhavee Public Health and Management. Healthc. Strateg. Plan. Soc. Incl. Dev. 2022, 1–40. [Google Scholar] [CrossRef]
- Volker, R.H.W.; Janis, A. Disease Transmission by Cannibalism: Rare Event or Common Occurrence? Proc. R. Soc. B Biol. Sci. 2007, 274, 1205–1210. [Google Scholar]
- Polis, G.A. The Evolution and Dynamics of Intraspecific Predation. Annu. Rev. Ecol. Syst. 2003, 12, 225–251. [Google Scholar] [CrossRef]
- Lindenbaum, S. Kuru Sorcery: Disease and Danger in the New Guinea Highlands; Routledge: Informa, UK, 2015; pp. 1–240. [Google Scholar]
- Matuschka, F.R.; Bannert, B. Recognition of Cyclic Transmission of Sarcocystis Stehlinii n. Sp. in the Gran Canarian Giant Lizard. J. Parasitol. 1989, 75, 383–387. [Google Scholar] [CrossRef] [PubMed]
- OIE. Mourilyan Virus. In OIE Aquatic Animal Disease Cards; OIE: Paris, France, 2007; pp. 1–2. [Google Scholar]
- OIE. Infection with Taura Syndrome Virus. In Manual of Diagnostic Tests for Aquatic Animals; OIE: Paris, France, 2019; Chapter 2.2.7; pp. 1–18. [Google Scholar]
- OIE. Infection with Yellow Head Virus Genotype 1. In Manual of Diagnostic Tests for Aquatic Animals; OIE: Paris, France, 2019; Chapter 2.2.9; pp. 1–14. [Google Scholar]
- OIE. Hepatopancreatic Parvovirus Disease. OIE Aquatic Animal Disease Cards. In Manual of Diagnostic Tests for Aquatic Animals; OIE: Paris, France, 2007; pp. 1–4. [Google Scholar]
- OIE. Infection with Macrobrachium Rosenbergii Nodavirus (White Tail Disease). In Manual of Diagnostic Tests for Aquatic Animals; OIE: Paris, France, 2019; Chapter 2.2.6; pp. 1–13. [Google Scholar]
- OIE. Infection with Necrotising Hepatopancreatitis. In Manual of Diagnostic Tests for Aquatic Animals; OIE: Paris, France, 2019; Chapter 2.2.3; pp. 1–12. [Google Scholar]
- OIE. Infection with Infectious Myonecrosis Virus. In Manual of Diagnostic Tests for Aquatic Animals; OIE: Paris, France, 2019; Chapter 2.2.5; pp. 2–3. [Google Scholar]
- OIE. Infection with Infectious Hypodermal and Haematopoietic Necrosis Virus. In Manual of Diagnostic Tests for Aquatic Animals; OIE: Paris, France, 2019; Chapter 2.2.4; pp. 1–18. [Google Scholar]
- OIE. Infection with White Spot Syndrome Virus. In Manual of Diagnostic Tests for Aquatic Animals; OIE: Paris, France, 2019; Chapter 2.2.8; pp. 1–16. [Google Scholar]
- Thuong, K. Van Experimental Studies on the Portal of Entry of White Spot Syndrome Virus in Penaeus Vannamei. Ph.D. Thesis, Ghent University, Faculty of Veterinary Medicine, Merelbeke, Belgium, 2016. [Google Scholar]
- Wang, X.; Chen, C.; Zhang, N.; Chen, Q.; Zhang, F.; Liu, X.; Li, F.; Shi, Z.-L.; Vlak, J.M.; Wang, M.; et al. Functional Peroral Infectivity Complex of White Spot Syndrome Virus of Shrimp. J. Virol. 2022, 96, e01173-22. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Yu, Z.; Song, X.; Guan, Y. Studies on the Transmission of WSSV (White Spot Syndrome Virus) in Juvenile Marsupenaeus Japonicus via Marine Microalgae. J. Invertebr. Pathol. 2007, 95, 87–92. [Google Scholar] [CrossRef] [PubMed]
- Jiang, G. Can White Spot Syndrome Virus Be Transmitted through the Phytoplankton→rotife →artemia→shrimp Pathway? Afr. J. Biotechnol. 2012, 11, 1277–1282. [Google Scholar] [CrossRef]
- Vanpatten, K.A.; Nunan, L.M.; Lightner, D.V. Seabirds as Potential Vectors of Penaeid Shrimp Viruses and the Development of a Surrogate Laboratory Model Utilizing Domestic Chickens. Aquaculture 2004, 241, 31–46. [Google Scholar] [CrossRef]
- Tang, K.F.J.; Pantoja, C.R.; Redman, R.M.; Lightner, D.V. A Histological Variant of White Spot Syndrome Virus (WSSV) from the Kingdom of Saudi Arabia. J. Invertebr. Pathol. 2013, 113, 82–85. [Google Scholar] [CrossRef] [PubMed]













| Group (n = 50) | Number of Shrimp/10 L | Number of Replicates | Probability of Infection |
|---|---|---|---|
| G10 | 10 | 5 | 1.00 a |
| G5 | 5 | 10 | 0.60 b |
| G3 | 3 | 17 | 0.59 b |
| G2 | 2 | 25 | 0.36 c |
| INDI | 1 | 40 | 0.18 d |
| Group | GI5h | GI48h | G∞ | INDI | GI72h |
|---|---|---|---|---|---|
| GI48h | 0.195 ns | - | - | - | - |
| G∞ | <0.001 * | 0.001 * | - | - | - |
| INDI | 0.116 ns | 0.004 * | <0.001 * | - | - |
| GI72h | 0.004 * | 0.111 ns | 0.204 ns | <0.001 * | - |
| GI30h | 0.578 ns | 0.064 ns | <0.001 * | 0.293 ns | <0.001 * |
| Group | № of Shrimp 10 L−1 | № of Replicates | № of Infected Populations/ Total № of Populations | Probability of Infection |
|---|---|---|---|---|
| GI5h_sentinel | 10 | 4 | 4/4 | 1.00 a |
| GI30h_sentinel | 10 | 4 | 0/4 | 0.00 b |
| GI48h_sentinel | 10 | 4 | 2/4 | 0.50 c |
| GI72h_sentinel | 10 | 4 | 3/4 | 0.75 c |
| Group (n = 50) | № of Shrimp 10 L−1 | № of Replicates | Environmental Component | № of Infected Populations/ Total № of Populations | Probability of Infection |
|---|---|---|---|---|---|
| G10-C | 10 | 5 | Molted cuticles | 0/5 | 0.00 a |
| G10-F | 10 | 5 | Feces | 1/5 | 0.20 b |
| G10-LT50-W | 10 | 5 | Water and biofilter material | 5/5 | 1.00 c |
| G10-LT50-SW | 10 | 5 | Water (no particles > 250 µm) | 4/5 | 0.80 c |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cox, N.; De Swaef, E.; Corteel, M.; Van Den Broeck, W.; Bossier, P.; Dantas-Lima, J.J.; Nauwynck, H.J. The Way of Water: Unravelling White Spot Syndrome Virus (WSSV) Transmission Dynamics in Litopenaeus vannamei Shrimp. Viruses 2023, 15, 1824. https://doi.org/10.3390/v15091824
Cox N, De Swaef E, Corteel M, Van Den Broeck W, Bossier P, Dantas-Lima JJ, Nauwynck HJ. The Way of Water: Unravelling White Spot Syndrome Virus (WSSV) Transmission Dynamics in Litopenaeus vannamei Shrimp. Viruses. 2023; 15(9):1824. https://doi.org/10.3390/v15091824
Chicago/Turabian StyleCox, Natasja, Evelien De Swaef, Mathias Corteel, Wim Van Den Broeck, Peter Bossier, João J. Dantas-Lima, and Hans J. Nauwynck. 2023. "The Way of Water: Unravelling White Spot Syndrome Virus (WSSV) Transmission Dynamics in Litopenaeus vannamei Shrimp" Viruses 15, no. 9: 1824. https://doi.org/10.3390/v15091824
APA StyleCox, N., De Swaef, E., Corteel, M., Van Den Broeck, W., Bossier, P., Dantas-Lima, J. J., & Nauwynck, H. J. (2023). The Way of Water: Unravelling White Spot Syndrome Virus (WSSV) Transmission Dynamics in Litopenaeus vannamei Shrimp. Viruses, 15(9), 1824. https://doi.org/10.3390/v15091824