Atomic Resolution Structure of the Oncolytic Parvovirus LuIII by Electron Microscopy and 3D Image Reconstruction
Abstract
:1. Introduction
2. Materials and Methods
2.1. LuIII VLP Expression and Purification
2.2. Sample Preparation and Cryo-Preservation
2.3. Cryo-EM Data Collection
2.4. Movie and Image Preprocessing, and 3D Map Reconstruction
2.5. Model Building and Structure Refinement
2.6. Structure Comparison
2.7. Figures
2.8. Accession Numbers
3. Results and Discussion
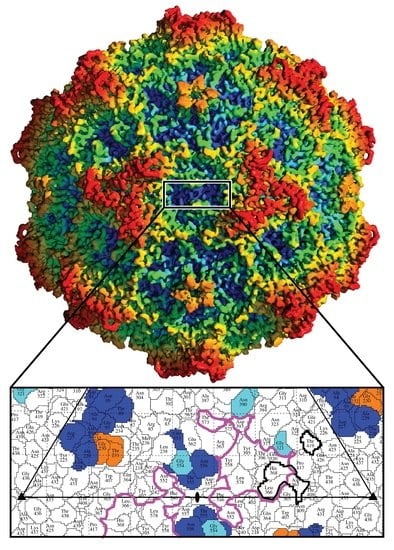
3.1. Cryo-EM and 3D Image Reconstruction Provides Atomic Resolution Information for LuIII VP2
3.2. LuIII Conserves General Parvoviral Structural Features
3.3. Variable Regions Confer Rodent Parvovirus Specific Surface Topologies
3.4. Variable Regions Confer Tumor Tropism and Cell Killing Efficiency
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Bell, J.; McFadden, G. Viruses for tumor therapy. Cell Host Microbe 2014, 15, 260–265. [Google Scholar] [CrossRef] [PubMed]
- Russell, S.J.; Peng, K.W.; Bell, J.C. Oncolytic virotherapy. Nat. Biotechnol. 2012, 30, 658–670. [Google Scholar] [CrossRef] [PubMed]
- Pol, J.; Buque, A.; Aranda, F.; Bloy, N.; Cremer, I.; Eggermont, A.; Erbs, P.; Fucikova, J.; Galon, J.; Limacher, J.M.; et al. Trial watch-oncolytic viruses and cancer therapy. Oncoimmunology 2016, 5, e1117740. [Google Scholar] [CrossRef] [PubMed]
- Greig, S.L. Talimogene laherparepvec: First global approval. Drugs 2016, 76, 147–154. [Google Scholar] [CrossRef] [PubMed]
- Rommelaere, J.; Geletneky, K.; Angelova, A.L.; Daeffler, L.; Dinsart, C.; Kiprianova, I.; Schlehofer, J.R.; Raykov, Z. Oncolytic parvoviruses as cancer therapeutics. Cytokine Growth Factor Rev. 2010, 21, 185–195. [Google Scholar] [CrossRef] [PubMed]
- Marchini, A.; Bonifati, S.; Scott, E.M.; Angelova, A.L.; Rommelaere, J. Oncolytic parvoviruses: From basic virology to clinical applications. Virol. J. 2015, 12, 6. [Google Scholar] [CrossRef] [PubMed]
- Paglino, J.; Tattersall, P. The parvoviral capsid controls an intracellular phase of infection essential for efficient killing of stepwise-transformed human fibroblasts. Virology 2011, 416, 32–41. [Google Scholar] [CrossRef] [PubMed]
- Guetta, E.; Graziani, Y.; Tal, J. Suppression of ehrlich ascites tumors in mice by minute virus of mice. J. Natl. Cancer Inst. 1986, 76, 1177–1180. [Google Scholar] [PubMed]
- Geletneky, K.; Kiprianova, I.; Ayache, A.; Koch, R.; Herrero, Y.C.M.; Deleu, L.; Sommer, C.; Thomas, N.; Rommelaere, J.; Schlehofer, J.R. Regression of advanced rat and human gliomas by local or systemic treatment with oncolytic parvovirus H-1 in rat models. Neuro-Oncology 2010, 12, 804–814. [Google Scholar] [CrossRef] [PubMed]
- Geletneky, K.; Nüesch, J.P.; Angelova, A.; Kiprianova, I.; Rommelaere, J. Double-faceted mechanism of parvoviral oncosuppression. Curr. Opin. Virol. 2015, 13, 17–24. [Google Scholar] [CrossRef] [PubMed]
- Geletneky, K.; Huesing, J.; Rommelaere, J.; Schlehofer, J.R.; Leuchs, B.; Dahm, M.; Krebs, O.; von Knebel Doeberitz, M.; Huber, B.; Hajda, J. Phase I/IIa study of intratumoral/intracerebral or intravenous/intracerebral administration of parvovirus H-1 (parvoryx) in patients with progressive primary or recurrent glioblastoma multiforme: ParvOryx01 protocol. BMC Cancer 2012, 12, 99. [Google Scholar] [CrossRef] [PubMed]
- Geletneky, K.; Hajda, J.; Angelova, A.L.; Leuchs, B.; Capper, D.; Bartsch, A.J.; Neumann, J.O.; Schoning, T.; Husing, J.; Beelte, B.; et al. Oncolytic H-1 parvovirus shows safety and signs of immunogenic activity in a first phase I/IIa glioblastoma trial. Mol. Ther. 2017. [Google Scholar] [CrossRef] [PubMed]
- Cotmore, S.F.; Agbandje-McKenna, M.; Chiorini, J.A.; Mukha, D.V.; Pintel, D.J.; Qiu, J.; Soderlund-Venermo, M.; Tattersall, P.; Tijssen, P.; Gatherer, D.; et al. The family parvoviridae. Arch. Virol. 2014, 159, 1239–1247. [Google Scholar] [CrossRef] [PubMed]
- Cotmore, S.F.; Tattersall, P. Structure and organization of the viral genome. In Parvoviruses; Hodder Arnold: London, UK, 2006; pp. 73–94. [Google Scholar]
- Richards, R.; Linser, P.; Armentrout, R.W. Kinetics of assembly of a parvovirus, minute virus of mice, in synchronized rat brain cells. J. Virol. 1977, 22, 778–793. [Google Scholar] [PubMed]
- Cotmore, S.F.; Tattersall, P. Parvoviruses: Small does not mean simple. Annu. Rev. Virol. 2013, 1, 517–537. [Google Scholar] [CrossRef] [PubMed]
- Angelova, A.L.; Geletneky, K.; Nuesch, J.P.; Rommelaere, J. Tumor selectivity of oncolytic parvoviruses: From in vitro and animal models to cancer patients. Front. Bioeng. Biotechnol. 2015, 3, 55. [Google Scholar] [CrossRef] [PubMed]
- Bar, S.; Rommelaere, J.; Nuesch, J.P. Pkη/Rdx-driven phosphorylation of PDK1: A novel mechanism promoting cancer cell survival and permissiveness for parvovirus-induced lysis. PLoS Pathog. 2015, 11, e1004703. [Google Scholar] [CrossRef] [PubMed]
- Zádori, Z.; Szelei, J.; Lacoste, M.C.; Li, Y.; Gariépy, S.; Raymond, P.; Allaire, M.; Nabi, I.R.; Tijssen, P. A viral phospholipase A2 is required for parvovirus infectivity. Dev. Cell 2001, 1, 291–302. [Google Scholar] [CrossRef]
- Tattersall, P.; Shatkin, A.J.; Ward, D.C. Sequence homology between the structural polypeptides of minute virus of mice. J. Mol. Biol. 1977, 111, 375–394. [Google Scholar] [CrossRef]
- Cotmore, S.F.; Tattersall, P. The autonomously replicating parvoviruses of vertebrates. Adv. Virus Res. 1987, 33, 91–174. [Google Scholar] [PubMed]
- Cotmore, S.F.; D’Abramo, A.M., Jr.; Ticknor, C.M.; Tattersall, P. Controlled conformational transitions in the mvm virion expose the vp1 n-terminus and viral genome without particle disassembly. Virology 1999, 254, 169–181. [Google Scholar] [CrossRef] [PubMed]
- Maroto, B.; Ramirez, J.C.; Almendral, J.M. Phosphorylation status of the parvovirus minute virus of mice particle: Mapping and biological relevance of the major phosphorylation sites. J. Virol. 2000, 74, 10892–10902. [Google Scholar] [CrossRef] [PubMed]
- Vollmers, E.M.; Tattersall, P. Distinct host cell fates for human malignant melanoma targeted by oncolytic rodent parvoviruses. Virology 2013, 446, 37–48. [Google Scholar] [CrossRef] [PubMed]
- Paglino, J.C.; Ozduman, K.; van den Pol, A.N. Luiii parvovirus selectively and efficiently targets, replicates in, and kills human glioma cells. J. Virol. 2012, 86, 7280–7291. [Google Scholar] [CrossRef] [PubMed]
- Cho, I.R.; Kaowinn, S.; Song, J.; Kim, S.; Koh, S.S.; Kang, H.Y.; Ha, N.C.; Lee, K.H.; Jun, H.S.; Chung, Y.H. VP2 capsid domain of the H-1 parvovirus determines susceptibility of human cancer cells to H-1 viral infection. Cancer Gene Ther. 2015, 22, 271–277. [Google Scholar] [CrossRef] [PubMed]
- Hernando, E.; Llamas-Saiz, A.L.; Foces-Foces, C.; McKenna, R.; Portman, I.; Agbandje-McKenna, M.; Almendral, J.M. Biochemical and physical characterization of parvovirus minute virus of mice virus-like particles. Virology 2000, 267, 299–309. [Google Scholar] [CrossRef] [PubMed]
- Kontou, M.; Govindasamy, L.; Nam, H.J.; Bryant, N.; Llamas-Saiz, A.L.; Foces-Foces, C.; Hernando, E.; Rubio, M.P.; McKenna, R.; Almendral, J.M.; et al. Structural determinants of tissue tropism and in vivo pathogenicity for the parvovirus minute virus of mice. J. Virol. 2005, 79, 10931–10943. [Google Scholar] [CrossRef] [PubMed]
- Halder, S.; Cotmore, S.; Heimburg-Molinaro, J.; Smith, D.F.; Cummings, R.D.; Chen, X.; Trollope, A.J.; North, S.J.; Haslam, S.M.; Dell, A.; et al. Profiling of glycan receptors for minute virus of mice in permissive cell lines towards understanding the mechanism of cell recognition. PLoS ONE 2014, 9, e86909. [Google Scholar] [CrossRef] [PubMed]
- Halder, S.; Nam, H.-J.; Govindasamy, L.; Vogel, M.; Dinsart, C.; Salomé, N.; McKenna, R.; Agbandje-McKenna, M. Structural characterization of H-1 parvovirus: Comparison of infectious virions to empty capsids. J. Virol. 2013, 87, 5128–5140. [Google Scholar] [CrossRef] [PubMed]
- Kailasan, S.; Garrison, J.; Ilyas, M.; Chipman, P.; McKenna, R.; Kantola, K.; Soderlund-Venermo, M.; Kucinskaite-Kodze, I.; Zvirbliene, A.; Agbandje-McKenna, M. Mapping antigenic epitopes on the human bocavirus capsid. J. Virol. 2016, 90, 4670–4680. [Google Scholar] [CrossRef] [PubMed]
- Potter, C.S.; Chu, H.; Frey, B.; Green, C.; Kisseberth, N.; Madden, T.J.; Miller, K.L.; Nahrstedt, K.; Pulokas, J.; Reilein, A.; et al. Leginon: A system for fully automated acquisition of 1000 electron micrographs a day. Ultramicroscopy 1999, 77, 153–161. [Google Scholar] [CrossRef]
- Zheng, S.Q.; Palovcak, E.; Armache, J.P.; Verba, K.A.; Cheng, Y.; Agard, D.A. Motioncor2: Anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 2017, 14, 331–332. [Google Scholar] [CrossRef] [PubMed]
- Rohou, A.; Grigorieff, N. CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 2015, 192, 216–221. [Google Scholar] [CrossRef] [PubMed]
- Yan, X.; Sinkovits, R.S.; Baker, T.S. AUTO3DEM—An automated and high throughput program for image reconstruction of icosahedral particles. J. Struct. Biol. 2007, 157, 73–82. [Google Scholar] [CrossRef] [PubMed]
- Emsley, P.; Cowtan, K. Coot: Model-building tools for molecular graphics. Acta Crystallogr. Sect. D Biol. Crystallogr. 2004, 60, 2126–2132. [Google Scholar] [CrossRef] [PubMed]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [PubMed]
- Bordoli, L.; Kiefer, F.; Arnold, K.; Benkert, P.; Battey, J.; Schwede, T. Protein structure homology modeling using SWISS-MODEL workspace. Nat. Protoc. 2008, 4, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Diffoot, N.; Chen, K.C.; Bates, R.C.; Lederman, M. The complete nucleotide sequence of parvovirus luiii and localization of a unique sequence possibly responsible for its encapsidation pattern. Virology 1993, 192, 339–345. [Google Scholar] [CrossRef] [PubMed]
- Carrillo-Tripp, M.; Shepherd, C.M.; Borelli, I.A.; Venkataraman, S.; Lander, G.; Natarajan, P.; Johnson, J.E.; Brooks, C.L., III; Reddy, V.S. VIPERdb2: An enhanced and web api enabled relational database for structural virology. Nucleic Acids Res. 2009, 37, D436–D442. [Google Scholar] [CrossRef] [PubMed]
- Tang, G.; Peng, L.; Baldwin, P.R.; Mann, D.S.; Jiang, W.; Rees, I.; Ludtke, S.J. EMAN2: An extensible image processing suite for electron microscopy. J. Struct. Biol. 2007, 157, 38–46. [Google Scholar] [CrossRef] [PubMed]
- Kleywegt, G.J.; Jones, T.A. XdlMAPMAN and xdlDATAMAN—Programs for reformatting, analysis and manipulation of biomacromolecular electron-density maps and reflection data sets. Acta Crystallogr. Sect. D Biol. Crystallogr. 1996, 52, 826–828. [Google Scholar] [CrossRef] [PubMed]
- Adams, P.D.; Afonine, P.V.; Bunkoczi, G.; Chen, V.B.; Davis, I.W.; Echols, N.; Headd, J.J.; Hung, L.W.; Kapral, G.J.; Grosse-Kunstleve, R.W.; et al. Phenix: A comprehensive python-based system for macromolecular structure solution. Acta Crystallogr. Sect. D Biol. Crystallogr. 2010, 66, 213–221. [Google Scholar] [CrossRef] [PubMed]
- Chen, V.B.; Arendall, W.B., III; Headd, J.J.; Keedy, D.A.; Immormino, R.M.; Kapral, G.J.; Murray, L.W.; Richardson, J.S.; Richardson, D.C. Molprobity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr. Sect. D Biol. Crystallogr. 2010, 66, 12–21. [Google Scholar] [CrossRef] [PubMed]
- Krissinel, E.; Henrick, K. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr. Sect. D Biol. Crystallogr. 2004, 60, 2256–2268. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Rossmann, M.G. The canine parvovirus empty capsid structure. J. Mol. Biol. 1993, 233, 231–244. [Google Scholar] [CrossRef] [PubMed]
- Simpson, A.A.; Hebert, B.; Sullivan, G.M.; Parrish, C.R.; Zadori, Z.; Tijssen, P.; Rossmann, M.G. The structure of porcine parvovirus: Comparison with related viruses. J. Mol. Biol. 2002, 315, 1189–1198. [Google Scholar] [CrossRef] [PubMed]
- Simpson, A.A.; Chandrasekar, V.; Hebert, B.; Sullivan, G.M.; Rossmann, M.G.; Parrish, C.R. Host range and variability of calcium binding by surface loops in the capsids of canine and feline parvoviruses. J. Mol. Biol. 2000, 300, 597–610. [Google Scholar] [CrossRef] [PubMed]
- Xiao, C.; Rossmann, M.G. Interpretation of electron density with stereographic roadmap projections. J. Struct. Biol. 2007, 158, 182–187. [Google Scholar] [CrossRef] [PubMed]
- Castellanos, M.; Perez, R.; Rodriguez-Huete, A.; Grueso, E.; Almendral, J.M.; Mateu, M.G. A slender tract of glycine residues is required for translocation of the VP2 protein n-terminal domain through the parvovirus mvm capsid channel to initiate infection. Biochem. J. 2013, 455, 87–94. [Google Scholar] [CrossRef] [PubMed]
- Halder, S.; Ng, R.; Agbandje-McKenna, M. Parvoviruses: Structure and infection. Future Virol. 2012, 7, 253–278. [Google Scholar] [CrossRef]
- Hafenstein, S.; Bowman, V.D.; Sun, T.; Nelson, C.D.; Palermo, L.M.; Chipman, P.R.; Battisti, A.J.; Parrish, C.R.; Rossmann, M.G. Structural comparison of different antibodies interacting with parvovirus capsids. J. Virol. 2009, 83, 5556–5566. [Google Scholar] [CrossRef] [PubMed]
- Kaufmann, B.; Lopez-Bueno, A.; Mateu, M.G.; Chipman, P.R.; Nelson, C.D.; Parrish, C.R.; Almendral, J.M.; Rossmann, M.G. Minute virus of mice, a parvovirus, in complex with the fab fragment of a neutralizing monoclonal antibody. J. Virol. 2007, 81, 9851–9858. [Google Scholar] [CrossRef] [PubMed]
- Wikoff, W.R.; Wang, G.; Parrish, C.R.; Cheng, R.H.; Strassheim, M.L.; Baker, T.S.; Rossmann, M.G. The structure of a neutralized virus: Canine parvovirus complexed with neutralizing antibody fragment. Structure 1994, 2, 595–607. [Google Scholar] [CrossRef]
- Lopez-Bueno, A.; Segovia, J.C.; Bueren, J.A.; O’Sullivan, M.G.; Wang, F.; Tattersall, P.; Almendral, J.M. Evolution to pathogenicity of the parvovirus minute virus of mice in immunodeficient mice involves genetic heterogeneity at the capsid domain that determines tropism. J. Virol. 2008, 82, 1195–1203. [Google Scholar] [CrossRef] [PubMed]
- Barbis, D.P.; Chang, S.F.; Parrish, C.R. Mutations adjacent to the dimple of the canine parvovirus capsid structure affect sialic acid binding. Virology 1992, 191, 301–308. [Google Scholar] [CrossRef]
- Lopez-Bueno, A.; Rubio, M.P.; Bryant, N.; McKenna, R.; Agbandje-McKenna, M.; Almendral, J.M. Host-selected amino acid changes at the sialic acid binding pocket of the parvovirus capsid modulate cell binding affinity and determine virulence. J. Virol. 2006, 80, 1563–1573. [Google Scholar] [CrossRef] [PubMed]
- Hueffer, K.; Govindasamy, L.; Agbandje-McKenna, M.; Parrish, C.R. Combinations of two capsid regions controlling canine host range determine canine transferrin receptor binding by canine and feline parvoviruses. J. Virol. 2003, 77, 10099–10105. [Google Scholar] [CrossRef] [PubMed]
- Govindasamy, L.; Hueffer, K.; Parrish, C.R.; Agbandje-McKenna, M. Structures of host range-controlling regions of the capsids of canine and feline parvoviruses and mutants. J. Virol. 2003, 77, 12211–12221. [Google Scholar] [CrossRef] [PubMed]
- Hafenstein, S.; Palermo, L.M.; Kostyuchenko, V.A.; Xiao, C.; Morais, M.C.; Nelson, C.D.; Bowman, V.D.; Battisti, A.J.; Chipman, P.R.; Parrish, C.R.; et al. Asymmetric binding of transferrin receptor to parvovirus capsids. Proc. Natl. Acad. Sci. USA 2007, 104, 6585–6589. [Google Scholar] [CrossRef] [PubMed]
- DiPrimio, N.; Asokan, A.; Govindasamy, L.; Agbandje-McKenna, M.; Samulski, R.J. Surface loop dynamics in adeno-associated virus capsid assembly. J. Virol. 2008, 82, 5178–5189. [Google Scholar] [CrossRef] [PubMed]
- Farr, G.A.; Tattersall, P. A conserved leucine that constricts the pore through the capsid fivefold cylinder plays a central role in parvoviral infection. Virology 2004, 323, 243–256. [Google Scholar] [CrossRef] [PubMed]
- Bleker, S.; Sonntag, F.; Kleinschmidt, J.A. Mutational analysis of narrow pores at the fivefold symmetry axes of adeno-associated virus type 2 capsids reveals a dual role in genome packaging and activation of phospholipase A2 activity. J. Virol. 2005, 79, 2528–2540. [Google Scholar] [CrossRef] [PubMed]
- Spalholz, B.A.; Tattersall, P. Interaction of minute virus of mice with differentiated cells: Strain-dependent target cell specificity is mediated by intracellular factors. J. Virol. 1983, 46, 937–943. [Google Scholar] [PubMed]
- Previsani, N.; Fontana, S.; Hirt, B.; Beard, P. Growth of the parvovirus minute virus of mice mvmp3 in el4 lymphocytes is restricted after cell entry and before viral dna amplification: Cell-specific differences in virus uncoating in vitro. J. Virol. 1997, 71, 7769–7780. [Google Scholar] [PubMed]
- Allaume, X.; El-Andaloussi, N.; Leuchs, B.; Bonifati, S.; Kulkarni, A.; Marttila, T.; Kaufmann, J.K.; Nettelbeck, D.M.; Kleinschmidt, J.; Rommelaere, J.; et al. Retargeting of rat parvovirus h-1pv to cancer cells through genetic engineering of the viral capsid. J. Virol. 2012, 86, 3452–3465. [Google Scholar] [CrossRef] [PubMed]
- Nam, H.J.; Gurda-Whitaker, B.; Gan, W.Y.; Ilaria, S.; McKenna, R.; Mehta, P.; Alvarez, R.A.; Agbandje-McKenna, M. Identification of the sialic acid structures recognized by minute virus of mice and the role of binding affinity in virulence adaptation. J. Biol. Chem. 2006, 281, 25670–25677. [Google Scholar] [CrossRef] [PubMed]
- Rubio, M.P.; Lopez-Bueno, A.; Almendral, J.M. Virulent variants emerging in mice infected with the apathogenic prototype strain of the parvovirus minute virus of mice exhibit a capsid with low avidity for a primary receptor. J. Virol. 2005, 79, 11280–11290. [Google Scholar] [CrossRef] [PubMed]
- Agbandje-McKenna, M.; Llamas-Saiz, A.L.; Wang, F.; Tattersall, P.; Rossmann, M.G. Functional implications of the structure of the murine parvovirus, minute virus of mice. Structure 1998, 6, 1369–1381. [Google Scholar] [CrossRef]
- Ball-Goodrich, L.J.; Tattersall, P. Two amino acid substitutions within the capsid are coordinately required for acquisition of fibrotropism by the lymphotropic strain of minute virus of mice. J. Virol. 1992, 66, 3415–3423. [Google Scholar] [PubMed]
- Ball-Goodrich, L.J.; Moir, R.D.; Tattersall, P. Parvoviral target cell specificity: Acquisition of fibrotropism by a mutant of the lymphotropic strain of minute virus of mice involves multiple amino acid substitutions within the capsid. Virology 1991, 184, 175–186. [Google Scholar] [CrossRef]
- Aydemir, F.; Salganik, M.; Resztak, J.; Singh, J.; Bennett, A.; Agbandje-McKenna, M.; Muzyczka, N. Mutants at the 2-fold interface of adeno-associated virus type 2 (AAV2) structural proteins suggest a role in viral transcription for aav capsids. J. Virol. 2016, 90, 7196–7204. [Google Scholar] [CrossRef] [PubMed]
- Salganik, M.; Aydemir, F.; Nam, H.J.; McKenna, R.; Agbandje-McKenna, M.; Muzyczka, N. Adeno-associated virus capsid proteins may play a role in transcription and second-strand synthesis of recombinant genomes. J. Virol. 2014, 88, 1071–1079. [Google Scholar] [CrossRef] [PubMed]







| Parameter | LuIII |
|---|---|
| Total No. of micrographs | 722 |
| Defocus range (μm) | 0.04–4.39 |
| Electron dose (e−/Å2) | 75 |
| No. of frames/micrograph | 50 |
| Pixel size (Å/pixel) | 1.064 |
| Starting No. of particles | 20,142 |
| No. of particles used for final map | 18,134 |
| Inverse B-factor used for final map (Å2) | 50 |
| Resolution of final map (Å) | 3.17 |
| Residue range | 37–587 |
| Map CC | 0.88 |
| RMSD (Å) | |
| Bonds | 0.0 |
| Angles | 0.8 |
| All-atom clash score | 11.8 |
| Ramachandran Plot (%) | |
| Favored | 97.8 |
| Allowed | 2.0 |
| Outliers | 0.2 |
| Rotamer outliers (%) | 0.2 |
| No. of Cβ deviations | 0.0 |
| Capsid Core | Non-Core β-Strands | ||
|---|---|---|---|
| Residue Range * | Secondary Structure | Residue Range # | VP Location |
| 125–133 | α-A | 82–86 | within BC loop |
| 51–54 | β-A | 105–109 | within BC loop |
| 211–213 | within EF loop | ||
| 61–74 | β-B | ||
| 521–535 | β-I | 156–158 | within DE loop |
| 137–147 | β-D | 165–167 | within DE loop |
| 268–272 | β-G | ||
| 374–378 | within GH loop | ||
| 112–115 | β-C | 393–396 | within GH loop |
| 496–500 | β-H | ||
| 176–179 | β-E | 345–346 | within GH loop |
| 255–258 | β-F | 353–354 | within GH loop |
| VR | Residue Range | RMSD (Å) | ||||
|---|---|---|---|---|---|---|
| LuIII | LuIII VP2 | MVM | H–1 | CPV | FPV | PPV |
| VR0 | 90(90–91)91 | 0.3–0.8 | 3.5–5.7 | 0.4–1.8 | 0.4–1.1 | 0.7–1.8 |
| VR0 | 92(none)94 | 0.6–1.2 | 0.6–1.1 | 1.3–1.9 | 1.0–1.6 | 2.1–2.7 |
| VR1 | 159(159–162)162 | 2.0–7.3 | 1.4–2.4 | 1.8–3.5 | 2.5–3.4 | 1.6–3.8 |
| VR2 | 227(227–230)230 | 0.8–3.1 | 2.1–3.5 | 1.7–1.8 | 0.6–3.0 | 1.5–2.2 |
| VR2 | 232(232–233)235 | 0.3–2.8 | 0.5–1.2 | 2.0–3.0 | 1.9–3.2 | 0.4–1.2 |
| VR2 | 237(none)241 | 0.3–1.0 | 0.2–0.7 | 1.8–3.2 | 1.7–3.1 | 2.0–5.0 |
| VR3 | 296(296–297)298 | 0.7–2.7 | 0.5–0.8 * | 1.4–3.3 | 1.7–3.8 | 2.2–4.1 |
| VR3 | 302(none)303 | 0.1–0.2 | 0.4 | 1.5–4.5 | 1.5–4.5 | 3.2–5.7 |
| VR4a | 310(none)312 | 0.5–0.6 | 0.3–0.5 | 2.5–3.4 | 1.7–2.4 | 0.5–1.2 |
| VR4a | 320(320)322 | 0.5–2.8 | 1.0–1.9 | 2.3–2.8 | 1.7–2.4 | 1.3–3.9 |
| VR5 | 359(none)367 | 0.3–1.2 | 0.2–1.6 | 2.0–6.8 | 0.9–6.0 | 0.6–1.6 |
| VR5 | 370(none)371 | 0.4–1.2 | 0.4–0.6 | 3.5–3.7 | 0.1–1.0 | 0.8–1.0 |
| VR6 | 386(390–391)392 | 0.7–4.7 | 0.6–3.2 | 2.7–4.6 | 0.9–4.5 | 1.3–3.5 |
| VR4b | 419(none)420 | 0.5–0.8 | 1.1–1.3 | 2.0–2.6 | 1.9–2.3 | 1.0–1.5 |
| VR7 | 511(511)512 | 0.8–0.9 | 1.9–2.0 | 1.8–2.8 | 2.2 | 1.4–1.5 |
| VR8 | 553(553–557)559 | 0.3–2.6 | 0.3–4.4 | 3.3–5.8 | 3.3–5.8 | 3.1–6.1 |
| Overall RMSD (LuIII) # | 0.7 | 0.7 | 1 | 1 | 1 | |
| % VP2 Identity (LuIII) $ | 73 | 70 | 53 | 52 | 52 | |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pittman, N.; Misseldine, A.; Geilen, L.; Halder, S.; Smith, J.K.; Kurian, J.; Chipman, P.; Janssen, M.; Mckenna, R.; Baker, T.S.; et al. Atomic Resolution Structure of the Oncolytic Parvovirus LuIII by Electron Microscopy and 3D Image Reconstruction. Viruses 2017, 9, 321. https://doi.org/10.3390/v9110321
Pittman N, Misseldine A, Geilen L, Halder S, Smith JK, Kurian J, Chipman P, Janssen M, Mckenna R, Baker TS, et al. Atomic Resolution Structure of the Oncolytic Parvovirus LuIII by Electron Microscopy and 3D Image Reconstruction. Viruses. 2017; 9(11):321. https://doi.org/10.3390/v9110321
Chicago/Turabian StylePittman, Nikéa, Adam Misseldine, Lorena Geilen, Sujata Halder, J. Kennon Smith, Justin Kurian, Paul Chipman, Mandy Janssen, Robert Mckenna, Timothy S. Baker, and et al. 2017. "Atomic Resolution Structure of the Oncolytic Parvovirus LuIII by Electron Microscopy and 3D Image Reconstruction" Viruses 9, no. 11: 321. https://doi.org/10.3390/v9110321
APA StylePittman, N., Misseldine, A., Geilen, L., Halder, S., Smith, J. K., Kurian, J., Chipman, P., Janssen, M., Mckenna, R., Baker, T. S., D’Abramo Jr., A., Cotmore, S., Tattersall, P., & Agbandje-McKenna, M. (2017). Atomic Resolution Structure of the Oncolytic Parvovirus LuIII by Electron Microscopy and 3D Image Reconstruction. Viruses, 9(11), 321. https://doi.org/10.3390/v9110321