Begomoviral Movement Protein Effects in Human and Plant Cells: Towards New Potential Interaction Partners
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Culture, Plants, and General Methods
2.2. Construction of Expression Plasmids for Cultured Cells
2.3. Construction of Expression Plasmids for Plants
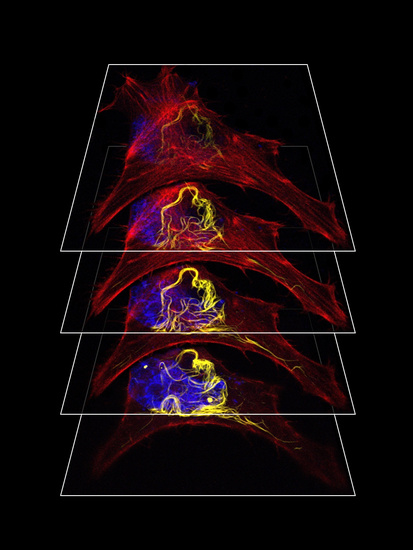
2.4. Transfection of Cultured Cell Lines and Immunofluorescence Analysis
2.5. Agro-Infiltration Assay and Microscopy
2.6. Immune-Affinity Capture of Proteins, Off-Bead Tryptic Digest, Nano-liquid chromatography (LC), and Tandem Mass Spectrometry (MS/MS) Analysis
2.7. Biolistic Inoculation of Plant Tissue
2.8. Tobacco Rattle Virus (TRV)-Based Virus-Induced Gene Silencing (VIGS) in N. benthamiana
2.9. Quantitative PCR (qPCR) Analysis
3. Results
3.1. Expression of MPAbMV in Mammalian Cell Lines Induced Filamentous Structures
3.2. Filamentous Structures Arise with Another Begomoviral MP as Well, but Not for an Unrelated Viral MP
3.3. MPAbMV Induces Re-Organization of Microtubules and Intermediate Filaments
3.4. MPAbMV Does Not Stabilize Microtubules
3.5. The MPAbMV Oligomerization Domain Is Dispensable for Interaction with the Microtubule Filaments
3.6. Expression of MPAbMV with and without Its Oligomerization Domain Has No Impact on the Microtubules Network in N. benthamiana Plant Cells
3.7. Examination of the MT Network in Triple Expression Experiments with AbMV MP, Rep, and MAP4:mCherry
3.8. Identification of Putative Novel MPAbMV Host Interaction Partners
3.9. Transient Silencing of Pin4 in N. benthamiana and Its Effect on AbMV
4. Discussion
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Anupam, V.; Malathi, V.G. Emerging geminivirus problems: A serious threat to crop production. Ann. Appl. Biol. 2003, 142, 145–164. [Google Scholar] [CrossRef]
- Zerbini, F.M.; Briddon, R.W.; Idris, A.; Martin, D.P.; Moriones, E.; Navas-castillo, J.; Rivera-bustamante, R.; Roumagnac, P.; Varsani, A.; Consortium, I.R. ICTV virus taxonomy profile: Dicistroviridae. J. Gen. Virol. 2017, 98, 355–356. [Google Scholar] [CrossRef]
- Jeske, H. Replication of geminiviruses and the use of rolling circle amplification for their diagnosis. In Tomato Yellow Leaf Curl Virus Disease; Czosnek, H., Ed.; Springer: Dordrecht, The Netherlands, 2007; pp. 141–156. [Google Scholar]
- Jeske, H. Geminiviruses. In Torque Teno Virus: The Still Elusive Human Pathogens; zur Hausen, H., de Villiers, E.-M., Eds.; Springer: Berlin, Germany, 2009; pp. 185–226. [Google Scholar]
- Hanley-Bowdoin, L.; Bejarano, E.R.; Robertson, D.; Mansoor, S. Geminiviruses: Masters at redirecting and reprogramming plant processes. Nat. Rev. Microbiol. 2013, 11, 777–788. [Google Scholar] [CrossRef] [PubMed]
- Horns, T.; Jeske, H. Localization of Abutilon Mosaic Virus (AbMV) DNA within leaf tissue by in situ hybridization. Virology 1991, 181, 580–588. [Google Scholar] [CrossRef]
- Wege, C.; Saunders, K.; Stanley, J.; Jeske, H. Comparative analysis of tissue tropism of bipartite geminiviruses. J. Phytopathol. 2001, 149, 359–368. [Google Scholar] [CrossRef]
- Zhang, S.C.; Wege, C.; Jeske, H. Movement proteins (BC1 and BV1) of Abutilon mosaic geminivirus are cotransported in and between cells of sink but not of source leaves as detected by green fluorescent protein tagging. Virology 2001, 290, 249–260. [Google Scholar] [CrossRef] [PubMed]
- Krenz, B.; Guo, T.W.; Kleinow, T. Stromuling when stressed! Acta Soc. Bot. Pol. 2014, 83, 325–329. [Google Scholar] [CrossRef]
- Krenz, B.; Windeisen, V.; Wege, C.; Jeske, H.; Kleinow, T. A plastid-targeted heat shock cognate 70 kDa protein interacts with the Abutilon mosaic virus movement protein. Virology 2010, 401, 6–17. [Google Scholar] [CrossRef] [PubMed]
- Krenz, B.; Jeske, H.; Kleinow, T. The induction of stromule formation by a plant DNA-virus in epidermal leaf tissues suggests a novel intra- and intercellular macromolecular trafficking route. Front. Plant Sci. 2012, 3, 291. [Google Scholar] [CrossRef] [PubMed]
- Kleinow, T.; Tanwir, F.; Kocher, C.; Krenz, B.; Wege, C.; Jeske, H. Expression dynamics and ultrastructural localization of epitope-tagged Abutilon mosaic virus nuclear shuttle and movement proteins in Nicotiana benthamiana cells. Virology 2009, 391, 212–220. [Google Scholar] [CrossRef] [PubMed]
- Hehnle, S.; Wege, C.; Jeske, H. Interaction of DNA with the movement proteins of geminiviruses revisited. J. Virol. 2004, 78, 7698–7706. [Google Scholar] [CrossRef] [PubMed]
- Frischmuth, S.; Kleinow, T.; Aberle, H.J.; Wege, C.; Hülser, D.; Jeske, H. Yeast two-hybrid systems confirm the membrane-association and oligomerization of BC1 but do not detect an interaction of the movement proteins BC1 and BV1 of Abutilon mosaic geminivirus. Arch. Virol. 2004, 149, 2349–2364. [Google Scholar] [CrossRef] [PubMed]
- Frischmuth, S.; Wege, C.; Hülser, D.; Jeske, H. The movement protein BC1 promotes redirection of the nuclear shuttle protein BV1 of Abutilon mosaic geminivirus to the plasma membrane in fission yeast. Protoplasma 2007, 230, 117–123. [Google Scholar] [CrossRef] [PubMed]
- Aberle, H.J.; Rütz, M.L.; Karayavuz, M.; Frischmuth, S.; Wege, C.; Hülser, D.; Jeske, H. Localizing the movement proteins of Abutilon mosaic geminivirus in yeast by subcellular fractionation and freeze-fracture immuno-labelling. Arch. Virol. 2002, 147, 103–117. [Google Scholar] [CrossRef]
- Rojas, M.R.; Hagen, C.; Lucas, W.J.; Gilbertson, R.L. Exploiting chinks in the plant’s armor: Evolution and emergence of geminiviruses. Annu. Rev. Phytopathol. 2005, 43, 361–394. [Google Scholar] [CrossRef] [PubMed]
- Rojas, M.R.; Noueiry, A.O.; Lucas, W.J.; Gilbertson, R.L. Bean dwarf mosaic geminivirus movement proteins recognize DNA in a form- and size-specific manner. Cell 1998, 95, 105–113. [Google Scholar] [CrossRef]
- Noueiry, A.O.; Lucas, W.J.; Gilbertson, R.L. Two proteins of a plant DNA virus coordinate nuclear and plasmodesmal transport. Cell 1994, 76, 925–932. [Google Scholar] [CrossRef]
- Levy, A.; Tzfira, T. Bean dwarf mosaic virus: A model system for the study of viral movement. Mol. Plant Pathol. 2010, 11, 451–461. [Google Scholar] [CrossRef] [PubMed]
- Levy, A.; Zheng, J.Y.; Lazarowitz, S.G. Synaptotagmin SYTA forms ER-Plasma membrane junctions that are recruited to plasmodesmata for plant virus movement. Curr. Biol. 2015, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Lewis, J.D.; Lazarowitz, S.G. Arabidopsis synaptotagmin SYTA regulates endocytosis and virus movement protein cell-to-cell transport. Proc. Natl. Acad. Sci. USA 2010, 107, 2491–2496. [Google Scholar] [CrossRef] [PubMed]
- Uchiyama, A.; Shimada-Beltran, H.; Levy, A.; Zheng, J.Y.; Javia, P.A.; Lazarowitz, S.G. The Arabidopsis synaptotagmin SYTA regulates the cell-to-cell movement of diverse plant viruses. Front. Plant Sci. 2014, 5, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Rojas, M.R.; Park, M.-R.; Seo, Y.-S.; Lucas, W.J.; Gilbertson, R.L. Histone H3 interacts and colocalizes with the nuclear shuttle protein and the movement protein of a geminivirus. J. Virol. 2011, 85, 11821–11832. [Google Scholar] [CrossRef] [PubMed]
- Dodding, M.P.; Way, M. Coupling viruses to dynein and kinesin-1. EMBO J. 2011, 30, 3527–3539. [Google Scholar] [CrossRef] [PubMed]
- Kodama, A.; Lechler, T.; Fuchs, E. Coordinating cytoskeletal tracks to polarize cellular movements. J. Cell Biol. 2004, 167, 203–207. [Google Scholar] [CrossRef] [PubMed]
- Radtke, K.; Döhner, K.; Sodeik, B. Viral interactions with the cytoskeleton: A hitchhiker’s guide to the cell. Cell. Microbiol. 2006, 8, 387–400. [Google Scholar] [CrossRef] [PubMed]
- Heinlein, M.; Padgett, H.S.; Gens, J.S.; Pickard, B.G.; Casper, S.J.; Epel, B.L.; Beachy, R.N. Changing patterns of localization of the tobacco mosaic virus movement protein and replicase to the endoplasmic reticulum and microtubules during infection. Plant Cell 1998, 10, 1107–1120. [Google Scholar] [CrossRef] [PubMed]
- Rojas, M.R.; Jiang, H.; Salati, R.; Xoconostle-Cázares, B.; Sudarshana, M.R.; Lucas, W.J.; Gilbertson, R.L. Functional analysis of proteins involved in movement of the monopartite begomovirus, Tomato yellow leaf curl virus. Virology 2001, 291, 110–125. [Google Scholar] [CrossRef] [PubMed]
- Ward, B.M.; Medville, R.; Lazarowitz, S.G.; Turgeon, R. The geminivirus BL1 movement protein is associated with endoplasmic reticulum-derived tubules in developing phloem cells. J. Virol. 1997, 71, 3726–3733. [Google Scholar] [PubMed]
- Heinlein, M. Plasmodesmata: Channels for viruses on the move. Methods Mol. Biol. 2015, 1217, 25–52. [Google Scholar] [CrossRef] [PubMed]
- Harries, P.A.; Schoelz, J.E.; Nelson, R.S. Intracellular transport of viruses and their components: Utilizing the cytoskeleton and membrane highways. Mol. Plant Microbe Interact. 2010, 23, 1381–1393. [Google Scholar] [CrossRef] [PubMed]
- Niehl, A.; Peña, E.J.; Amari, K.; Heinlein, M. Microtubules in viral replication and transport. Plant J. 2013, 75, 290–308. [Google Scholar] [CrossRef] [PubMed]
- Boyko, V.; Ferralli, J.; Ashby, J.; Schellenbaum, P.; Heinlein, M. Function of microtubules in intercellular transport of plant virus RNA. Nat. Cell Biol. 2000, 2, 826–832. [Google Scholar] [CrossRef] [PubMed]
- Boyko, V.; Ferralli, J.; Heinlein, M. Cell-to-cell movement of TMV RNA is temperature-dependent and corresponds to the association of movement protein with microtubules. Plant J. 2000, 22, 315–325. [Google Scholar] [CrossRef] [PubMed]
- Ferralli, J.; Ashby, J.; Fasler, M.; Boyko, V.; Heinlein, M. Disruption of microtubule organization and centrosome function by expression of tobacco mosaic virus movement protein. J. Virol. 2006, 80, 5807–5821. [Google Scholar] [CrossRef] [PubMed]
- Wileman, T. Aggresomes and pericentriolar sites of virus assembly: Cellular defense or viral design? Annu. Rev. Microbiol. 2007, 61, 149–167. [Google Scholar] [CrossRef] [PubMed]
- Sanderfoot, A.A.; Lazarowitz, S.G. Cooperation in Viral Movement: The geminivirus BL1 movement protein interacts with br1 and redirects it from the nucleus to the cell periphery. Plant Cell 1995, 7, 1185–1194. [Google Scholar] [CrossRef] [PubMed]
- Krenz, B.; Schießl, I.; Greiner, E.; Krapp, S. Analyses of pea necrotic yellow dwarf virus-encoded proteins. Virus Genes 2017, 53, 454–463. [Google Scholar] [CrossRef] [PubMed]
- Striebinger, H.; Funk, C.; Raschbichler, V.; Bailer, S.M. Subcellular trafficking and functional relationship of the HSV-1 glycoproteins N and M. Viruses 2016, 8, 83. [Google Scholar] [CrossRef] [PubMed]
- Krapp, S.; Greiner, E.; Amin, B.; Sonnewald, U.; Krenz, B. The stress granule component G3BP is a novel interaction partner for the nuclear shuttle proteins of the nanovirus pea necrotic yellow dwarf virus and geminivirus abutilon mosaic virus. Virus Res. 2017, 227, 6–14. [Google Scholar] [CrossRef] [PubMed]
- Noris, E.; Miozzi, L. Real-time PCR protocols for the quantification of the begomovirus tomato yellow leaf curl sardinia virus in tomato plants and in its insect vector. Methods Mol. Biol. 2015, 1236, 61–72. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.; Fernald, R.D. Comprehensive algorithm for quantitative real-time polymerase chain reaction. J. Comput. Biol. 2005, 12, 1047–1064. [Google Scholar] [CrossRef] [PubMed]
- Vandesompele, J.; de Preter, K.; Pattyn, I.; Poppe, B.; van Roy, N.; de Paepe, A.; Speleman, R. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002, 3, 34–41. [Google Scholar] [CrossRef] [Green Version]
- Martinière, A.; Gargani, D.; Uzest, M.; Lautredou, N.; Blanc, S.; Drucker, M. A role for plant microtubules in the formation of transmission-specific inclusion bodies of Cauliflower mosaic virus. Plant J. 2009, 58, 135–146. [Google Scholar] [CrossRef] [PubMed]
- Wege, C.; Pohl, D. Abutilon mosaic virus DNA B component supports mechanical virus transmission, but does not counteract begomoviral phloem limitation in transgenic plants. Virology 2007, 365, 173–186. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.C.; Ghosh, R.; Jeske, H. Subcellular targeting domains of Abutilon mosaic geminivirus movement protein BC1. Arch. Virol. 2002, 147, 2349–2363. [Google Scholar] [CrossRef] [PubMed]
- Ratcliff, F.; Baulcombe, D.C.; Lane, C.; Nr, N.; Martin-Hernandez, A.M.; Baulcombe, D.C. Tobacco rattle virus as a vector for analysis of gene function by silencing. Science 2001, 25, 237–245. [Google Scholar]
- Liu, Y.L.; Schiff, M.; Dinesh-Kumar, S.P. Virus-induced gene silencing in tomato. Plant J. 2002, 31, 777–786. [Google Scholar] [CrossRef] [PubMed]
- Herrmann, H.; Bär, H.; Kreplak, L.; Strelkov, S.V.; Aebi, U. Intermediate filaments: From cell architecture to nanomechanics. Mol. Cell Biol. 2007, 8, 562–573. [Google Scholar] [CrossRef] [PubMed]
- Gruenbaum, Y.; Margalit, A.; Goldman, R.D.; Shumaker, D.K.; Wilson, K.L. The nuclear lamina comes of age. Nat. Rev. Mol. Cell Biol. 2005, 6, 21–31. [Google Scholar] [CrossRef] [PubMed]
- Goldman, R.D.; Khuon, S.; Chou, Y.H.; Opal, P.; Steinert, P.M. The function of intermediate filaments in cell shape and cytoskeletal integrity. J. Cell Biol. 1996, 134, 971–983. [Google Scholar] [CrossRef] [PubMed]
- Herrmann, H.; Strelkov, S.V. History and phylogeny of intermediate filaments: Now in insects. BMC Biol. 2011, 9, 16. [Google Scholar] [CrossRef] [PubMed]
- Mizuno, K. A cytoskeletal 50 kDa protein in higher plants that forms intermediate-sized filaments and stabilizes microtubules. Protoplasma 1995, 186, 99–112. [Google Scholar] [CrossRef]
- Goodbody, K.I.M.C.; Hargreaves, A.J.; Lloyd, C.W. On the distribution of microtubule-associated intermediate filament antigens in plant suspension cells. J. Cell Sci. 1989, 93, 427–438. [Google Scholar]
- Pryer, N.K.; Walker, R.A.; Skeen, V.P.; Bourns, B.D.; Soboeiro, M.F.; Salmon, E.D. Brain microtubule-associated proteins modulate microtubule dynamic instability in vitro. Real-time observations using video microscopy. J. Cell Sci. 1992, 103, 965–976. [Google Scholar] [PubMed]
- Rutten, T.; Chan, J.; Lloyd, C.W. A 60-kDa plant microtubule-associated protein promotes the growth and stabilization of neurotubules in vitro. Proc. Natl. Acad. Sci. USA 1997, 94, 4469–4474. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, H.L.; Chari, S.; Gruber, D.; Lue, C.M.; Chapin, S.J.; Bulinski, J.C. Overexpression of full- or partial-length MAP4 stabilizes microtubules and alters cell growth. J. Cell Sci. 1997, 110, 281–294. [Google Scholar] [PubMed]
- McMichael, C.M.; Reynolds, G.D.; Koch, L.M.; Wang, C.; Jiang, N.; Nadeau, J.; Sack, F.D.; Gelderman, M.B.; Pan, J.; Bednarek, S.Y. Mediation of clathrin-dependent trafficking during cytokinesis and cell expansion by arabidopsis stomatal cytokinesis defective proteins. Plant Cell 2013, 25, 3910–3925. [Google Scholar] [CrossRef] [PubMed]
- Fujiyama-Nakamura, S.; Yoshikawa, H.; Homma, K.; Hayano, T.; Tsujimura-Takahashi, T.; Izumikawa, K.; Ishikawa, H.; Miyazawa, N.; Yanagida, M.; Miura, Y.; et al. Parvulin (Par14), a peptidyl-prolyl cis-trans isomerase, is a novel rRNA processing factor that evolved in the metazoan lineage. Mol. Cell. Proteom. 2009, 8, 1552–1565. [Google Scholar] [CrossRef] [PubMed]
- Thiele, A.; Krentzlin, K.; Erdmann, F.; Rauh, D.; Hause, G.; Zerweck, J.; Kilka, S.; Pösel, S.; Fischer, G.; Schutkowski, M.; et al. Parvulin 17 promotes microtubule assembly by its peptidyl-prolyl cis/trans isomerase activity. J. Mol. Biol. 2011, 411, 896–909. [Google Scholar] [CrossRef] [PubMed]
- Burgardt, N.I.; Schmidt, A.; Manns, A.; Schutkowski, A.; Jahreis, G.; Lin, Y.J.; Schulze, B.; Masch, A.; Lücke, C.; Weiwad, M. Parvulin 17-catalyzed tubulin polymerization is regulated by calmodulin in a calcium-dependent manner. J. Biol. Chem. 2015, 290, 16708–16722. [Google Scholar] [CrossRef] [PubMed]
- Saningong, A.D.; Bayer, P. Human DNA-binding peptidyl-prolyl cis/trans isomerase Par14 is cell cycle dependently expressed and associates with chromatin in vivo. BMC Biochem. 2015, 16, 4. [Google Scholar] [CrossRef] [PubMed]
- Ascencio-Ibanez, J.T.; Sozzani, R.; Lee, T.-J.; Chu, T.-M.; Wolfinger, R.D.; Cella, R.; Hanley-Bowdoin, L. Global analysis of arabidopsis gene expression uncovers a complex array of changes impacting pathogen response and cell cycle during geminivirus infection. Plant Physiol. 2008, 148, 436–454. [Google Scholar] [CrossRef] [PubMed]
- Milbradt, J.; Hutterer, C.; Bahsi, H.; Wagner, S.; Sonntag, E.; Horn, A.H.C.; Kaufer, B.B.; Mori, Y.; Sticht, H.; Fossen, T.; et al. The prolyl isomerase Pin1 promotes the herpesvirus-induced phosphorylation-dependent disassembly of the nuclear lamina required for nucleocytoplasmic egress. PLoS Pathog. 2016, 12, 1–30. [Google Scholar] [CrossRef] [PubMed]








© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Krapp, S.; Schuy, C.; Greiner, E.; Stephan, I.; Alberter, B.; Funk, C.; Marschall, M.; Wege, C.; Bailer, S.M.; Kleinow, T.; et al. Begomoviral Movement Protein Effects in Human and Plant Cells: Towards New Potential Interaction Partners. Viruses 2017, 9, 334. https://doi.org/10.3390/v9110334
Krapp S, Schuy C, Greiner E, Stephan I, Alberter B, Funk C, Marschall M, Wege C, Bailer SM, Kleinow T, et al. Begomoviral Movement Protein Effects in Human and Plant Cells: Towards New Potential Interaction Partners. Viruses. 2017; 9(11):334. https://doi.org/10.3390/v9110334
Chicago/Turabian StyleKrapp, Susanna, Christian Schuy, Eva Greiner, Irina Stephan, Barbara Alberter, Christina Funk, Manfred Marschall, Christina Wege, Susanne M. Bailer, Tatjana Kleinow, and et al. 2017. "Begomoviral Movement Protein Effects in Human and Plant Cells: Towards New Potential Interaction Partners" Viruses 9, no. 11: 334. https://doi.org/10.3390/v9110334
APA StyleKrapp, S., Schuy, C., Greiner, E., Stephan, I., Alberter, B., Funk, C., Marschall, M., Wege, C., Bailer, S. M., Kleinow, T., & Krenz, B. (2017). Begomoviral Movement Protein Effects in Human and Plant Cells: Towards New Potential Interaction Partners. Viruses, 9(11), 334. https://doi.org/10.3390/v9110334