Recognizing the SINEs of Infection: Regulation of Retrotransposon Expression and Modulation of Host Cell Processes
Abstract
:1. Introduction
2. Structure and Diversity of SINEs
3. Transcriptional Regulation of SINEs
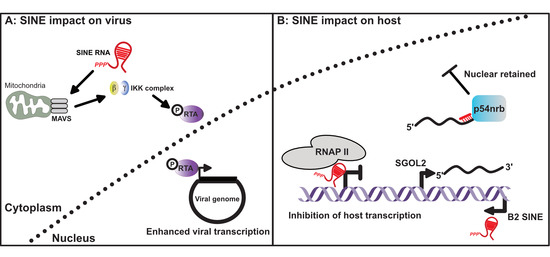
4. Induction and Consequences of SINE Expression
5. Closing Remarks
Conflicts of Interest
References
- Lander, E.S.; Linton, L.M.; Birren, B.; Nusbaum, C.; Zody, M.C.; Baldwin, J.; Devon, K.; Dewar, K.; Doyle, M.; FitzHugh, W.; et al. Initial sequencing and analysis of the human genome. Nature 2001, 409, 860–921. [Google Scholar] [CrossRef] [PubMed]
- Venter, J.C.; Adams, M.D.; Myers, E.W.; Li, P.W.; Mural, R.J.; Sutton, G.G.; Smith, H.O.; Yandell, M.; Evans, C.A.; Holt, R.A.; et al. The sequence of the human genome. Science 2001, 291, 1304–1351. [Google Scholar] [CrossRef] [PubMed]
- Wessler, S.R. Transposable elements and the evolution of eukaryotic genomes. Proc. Natl. Acad. Sci. USA 2006, 103, 17600–17601. [Google Scholar] [CrossRef] [PubMed]
- Cordaux, R.; Batzer, M.A. The impact of retrotransposons on human genome evolution. Nat. Rev. Genet. 2009, 10, 691–703. [Google Scholar] [CrossRef] [PubMed]
- Hayward, A.; Katzourakis, A. Endogenous retroviruses. Curr. Biol. 2015, 25, R644–R646. [Google Scholar] [CrossRef] [PubMed]
- Daniels, G.R.; Deininger, P.L. Repeat sequence families derived from mammalian tRNA genes. Nature 1985, 317, 819–822. [Google Scholar] [CrossRef] [PubMed]
- Kriegs, J.O.; Churakov, G.; Jurka, J.; Brosius, J.; Schmitz, J. Evolutionary history of 7SL RNA-derived SINEs in Supraprimates. Trends Genet. 2007, 23, 158–161. [Google Scholar] [CrossRef] [PubMed]
- Ullu, E.; Tschudi, C. Alu sequences are processed 7SL RNA genes. Nature 1984, 312, 171–172. [Google Scholar] [CrossRef] [PubMed]
- Weiner, A.M. An abundant cytoplasmic 7S RNA is complementary to the dominant interspersed middle repetitive DNA sequence family in the human genome. Cell 1980, 22, 209–218. [Google Scholar] [CrossRef]
- Deininger, P. Alu elements: Know the SINEs. Genome Biol. 2011, 12, 236. [Google Scholar] [CrossRef] [PubMed]
- Feng, Q.; Moran, J.V.; Kazazian, H.H.; Boeke, J.D. Human L1 retrotransposon encodes a conserved endonuclease required for retrotransposition. Cell 1996, 87, 905–916. [Google Scholar] [CrossRef]
- Martin, S.L.; Li, J.; Epperson, L.E.; Lieberman, B. Functional reverse transcriptases encoded by A-type mouse LINE-1: Defining the minimal domain by deletion analysis. Gene 1998, 215, 69–75. [Google Scholar] [CrossRef]
- Moran, J.V.; Holmes, S.E.; Naas, T.P.; DeBerardinis, R.J.; Boeke, J.D.; Kazazian, H.H. High frequency retrotransposition in cultured mammalian cells. Cell 1996, 87, 917–927. [Google Scholar] [CrossRef]
- Slotkin, R.K.; Martienssen, R. Transposable elements and the epigenetic regulation of the genome. Nat. Rev. Genet. 2007, 8, 272–285. [Google Scholar] [CrossRef] [PubMed]
- McCarthy, E.M.; McDonald, J.F. Long terminal repeat retrotransposons of Mus musculus. Genome Biol. 2004, 5, R14. [Google Scholar] [CrossRef] [PubMed]
- Mouse Genome Sequencing Consortium; Waterston, R.H.; Lindblad-Toh, K.; Birney, E.; Rogers, J.; Abril, J.F.; Agarwal, P.; Agarwala, R.; Ainscough, R.; Alexandersson, M.; et al. Initial sequencing and comparative analysis of the mouse genome. Nature 2002, 420, 520–562. [Google Scholar] [CrossRef]
- Krayev, A.S.; Kramerov, D.A.; Skryabin, K.G.; Ryskov, A.P.; Bayev, A.A.; Georgiev, G.P. The nucleotide sequence of the ubiquitous repetitive DNA sequence B1 complementary to the most abundant class of mouse fold-back RNA. Nucleic Acids Res. 1980, 8, 1201–1215. [Google Scholar] [CrossRef] [PubMed]
- Abdurashitov, M.A.; Tomilov, V.N.; Chernukhin, V.A.; Degtyarev, S.K. A physical map of human Alu repeats cleavage by restriction endonucleases. BMC Genom. 2008, 9, 305. [Google Scholar] [CrossRef] [PubMed]
- Deininger, P.L.; Moran, J.V.; Batzer, M.A.; Kazazian, H.H. Mobile elements and mammalian genome evolution. Curr. Opin. Genet. Dev. 2003, 13, 651–658. [Google Scholar] [CrossRef] [PubMed]
- Kramerov, D.A.; Vassetzky, N.S. SINEs. Wiley Interdiscip. Rev. RNA 2011, 2, 772–786. [Google Scholar] [CrossRef] [PubMed]
- Burke, J.M.; Sullivan, C.S. DUSP11—An RNA phosphatase that regulates host and viral non-coding RNAs in mammalian cells. RNA Biol. 2017, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Burke, J.M.; Kincaid, R.P.; Nottingham, R.M.; Lambowitz, A.M.; Sullivan, C.S. DUSP11 activity on triphosphorylated transcripts promotes Argonaute association with noncanonical viral microRNAs and regulates steady-state levels of cellular noncoding RNAs. Genes Dev. 2016, 30, 2076–2092. [Google Scholar] [CrossRef] [PubMed]
- Shumyatsky, G.; Wright, D.; Reddy, R. Methylphosphate cap structure increases the stability of 7SK, B2 and U6 small RNAs in Xenopus oocytes. Nucleic Acids Res. 1993, 21, 4756–4761. [Google Scholar] [CrossRef] [PubMed]
- Dewannieux, M.; Heidmann, T. Role of poly(A) tail length in Alu retrotransposition. Genomics 2005, 86, 378–381. [Google Scholar] [CrossRef] [PubMed]
- Borodulina, O.R.; Kramerov, D.A. Transcripts synthesized by RNA polymerase III can be polyadenylated in an AAUAAA-dependent manner. RNA 2008, 14, 1865–1873. [Google Scholar] [CrossRef] [PubMed]
- Paule, M.R.; White, R.J. Transcription by RNA polymerases I and III. Nucleic Acids Res. 2000, 28, 1283–1298. [Google Scholar] [CrossRef] [PubMed]
- Schramm, L.; Hernandez, N. Recruitment of RNA polymerase III to its target promoters. Genes Dev. 2002, 16, 2593–2620. [Google Scholar] [CrossRef] [PubMed]
- Arimbasseri, A.G.; Maraia, R.J. RNA polymerase III advances: Structural and tRNA functional views. Trends Biochem. Sci. 2016, 41, 546–559. [Google Scholar] [CrossRef] [PubMed]
- Sharp, S.; DeFranco, D.; Dingermann, T.; Farrell, P.; Söll, D. Internal control regions for transcription of eukaryotic tRNA genes. Proc. Natl. Acad. Sci. USA 1981, 78, 6657–6661. [Google Scholar] [CrossRef] [PubMed]
- Galli, G.; Hofstetter, H.; Birnstiel, M.L. Two conserved sequence blocks within eukaryotic tRNA genes are major promoter elements. Nature 1981, 294, 626–631. [Google Scholar] [CrossRef] [PubMed]
- Lassar, A.B.; Martin, P.L.; Roeder, R.G. Transcription of class III genes: Formation of preinitiation complexes. Science 1983, 222, 740–748. [Google Scholar] [CrossRef] [PubMed]
- Bieker, J.J.; Martin, P.L.; Roeder, R.G. Formation of a rate-limiting intermediate in 5S RNA gene transcription. Cell 1985, 40, 119–127. [Google Scholar] [CrossRef]
- Setzer, D.R.; Brown, D.D. Formation and stability of the 5 S RNA transcription complex. J. Biol. Chem. 1985, 260, 2483–2492. [Google Scholar] [PubMed]
- Nielsen, S.; Yuzenkova, Y.; Zenkin, N. Mechanism of eukaryotic RNA polymerase III transcription termination. Science 2013, 340, 1577–1580. [Google Scholar] [CrossRef] [PubMed]
- Matsuzaki, H.; Kassavetis, G.A.; Geiduschek, E.P. Analysis of RNA chain elongation and termination by Saccharomyces cerevisiae RNA polymerase III. J. Mol. Biol. 1994, 235, 1173–1192. [Google Scholar] [CrossRef] [PubMed]
- Bogenhagen, D.F.; Brown, D.D. Nucleotide sequences in Xenopus 5S DNA required for transcription termination. Cell 1981, 24, 261–270. [Google Scholar] [CrossRef]
- Orioli, A.; Pascali, C.; Pagano, A.; Teichmann, M.; Dieci, G. RNA polymerase III transcription control elements: Themes and variations. Gene 2012, 493, 185–194. [Google Scholar] [CrossRef] [PubMed]
- Braglia, P.; Percudani, R.; Dieci, G. Sequence context effects on oligo(dT) termination signal recognition by Saccharomyces cerevisiae RNA polymerase III. J. Biol. Chem. 2005, 280, 19551–19562. [Google Scholar] [CrossRef] [PubMed]
- Siegfried, Z.; Simon, I. DNA methylation and gene expression. Wiley Interdiscip. Rev. Syst. Biol. Med. 2010, 2, 362–371. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.M.; Maraia, R.J.; Rubin, C.M.; Schmid, C.W. Alu transcripts: Cytoplasmic localisation and regulation by DNA methylation. Nucleic Acids Res. 1994, 22, 1087–1095. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.-M.; Schmid, C.W. Proposed roles for DNA methylation in Alu transcriptional repression and mutational inactivation. Nucleic Acids Res. 1993, 21, 1351–1359. [Google Scholar] [CrossRef] [PubMed]
- Xie, H.; Wang, M.; Bonaldo, M.F.; Rajaram, V.; Stellpflug, W.; Smith, C.; Arndt, K.; Goldman, S.; Tomita, T.; Soares, M.B. Epigenomic analysis of Alu repeats in human ependymomas. Proc. Natl. Acad. Sci. USA 2010, 107, 6952–6957. [Google Scholar] [CrossRef] [PubMed]
- Varshney, D.; Vavrova-Anderson, J.; Oler, A.J.; Cowling, V.H.; Cairns, B.R.; White, R.J. SINE transcription by RNA polymerase III is suppressed by histone methylation but not by DNA methylation. Nat. Commun. 2015, 6, 6569. [Google Scholar] [CrossRef] [PubMed]
- Martens, J.H.A.; O’Sullivan, R.J.; Braunschweig, U.; Opravil, S.; Radolf, M.; Steinlein, P.; Jenuwein, T. The profile of repeat-associated histone lysine methylation states in the mouse epigenome. EMBO J. 2005, 24, 800–812. [Google Scholar] [CrossRef] [PubMed]
- Muramoto, H.; Yagi, S.; Hirabayashi, K.; Sato, S.; Ohgane, J.; Tanaka, S.; Shiota, K. Enrichment of short interspersed transposable elements to embryonic stem cell-specific hypomethylated gene regions. Genes Cells 2010, 15, 855–865. [Google Scholar] [CrossRef] [PubMed]
- Román, A.C.; González-Rico, F.J.; Fernández-Salguero, P.M. B1-SINE retrotransposons. Mob. Genet. Elem. 2011, 1, 66–70. [Google Scholar] [CrossRef] [PubMed]
- Ichiyanagi, K.; Li, Y.; Watanabe, T.; Ichiyanagi, T.; Fukuda, K.; Kitayama, J.; Yamamoto, Y.; Kuramochi-Miyagawa, S.; Nakano, T.; Yabuta, Y.; et al. Locus- and domain-dependent control of DNA methylation at mouse B1 retrotransposons during male germ cell development. Genome Res. 2011, 21, 2058–2066. [Google Scholar] [CrossRef] [PubMed]
- Bachvarova, R. Small B2 RNAs in mouse oocytes, embryos, and somatic tissues. Dev. Biol. 1988, 130, 513–523. [Google Scholar] [CrossRef]
- Williams, B.R. PKR; a sentinel kinase for cellular stress. Oncogene 1999, 18, 6112–6120. [Google Scholar] [CrossRef] [PubMed]
- Chu, W.M.; Ballard, R.; Carpick, B.W.; Williams, B.R.; Schmid, C.W. Potential Alu function: Regulation of the activity of double-stranded RNA-activated kinase PKR. Mol. Cell. Biol. 1998, 18, 58–68. [Google Scholar] [CrossRef] [PubMed]
- Mariner, P.D.; Walters, R.D.; Espinoza, C.A.; Drullinger, L.F.; Wagner, S.D.; Kugel, J.F.; Goodrich, J.A. Human Alu RNA is a modular transacting repressor of mRNA transcription during heat shock. Mol. Cell 2008, 29, 499–509. [Google Scholar] [CrossRef] [PubMed]
- Fornace, A.J.; Mitchell, J.B. Induction of B2 RNA polymerase III transcription by heat shock: Enrichment for heat shock induced sequences in rodent cells by hybridization subtraction. Nucleic Acids Res. 1986, 14, 5793–5811. [Google Scholar] [CrossRef] [PubMed]
- Espinoza, C.A.; Allen, T.A.; Hieb, A.R.; Kugel, J.F.; Goodrich, J.A. B2 RNA binds directly to RNA polymerase II to repress transcript synthesis. Nat. Struct. Mol. Biol. 2004, 11, 822–829. [Google Scholar] [CrossRef] [PubMed]
- Zovoilis, A.; Cifuentes-Rojas, C.; Chu, H.-P.; Hernandez, A.J.; Lee, J.T. Destabilization of B2 RNA by EZH2 Activates the Stress Response. Cell 2016, 167, 1788–1802.e13. [Google Scholar] [CrossRef] [PubMed]
- Panning, B.; Smiley, J.R. Activation of RNA polymerase III transcription of human Alu repetitive elements by adenovirus type 5: Requirement for the E1b 58-kilodalton protein and the products of E4 open reading frames 3 and 6. Mol. Cell. Biol. 1993, 13, 3231–3244. [Google Scholar] [CrossRef] [PubMed]
- Williams, W.P.; Tamburic, L.; Astell, C.R. Increased levels of B1 and B2 SINE transcripts in mouse fibroblast cells due to minute virus of mice infection. Virology 2004, 327, 233–241. [Google Scholar] [CrossRef] [PubMed]
- Singh, K.; Carey, M.; Saragosti, S.; Botchan, M. Expression of enhanced levels of small RNA polymerase III transcripts encoded by the B2 repeats in simian virus 40-transformed mouse cells. Nature 1985, 314, 553–556. [Google Scholar] [CrossRef] [PubMed]
- Panning, B.; Smiley, J.R. Activation of RNA polymerase III transcription of human Alu elements by herpes simplex virus. Virology 1994, 202, 408–417. [Google Scholar] [CrossRef] [PubMed]
- Jang, K.L.; Latchman, D.S. The herpes simplex virus immediate-early protein ICP27 stimulates the transcription of cellular Alu repeated sequences by increasing the activity of transcription factor TFIIIC. Biochem. J. 1992, 284, 667–673. [Google Scholar] [CrossRef] [PubMed]
- Karijolich, J.; Abernathy, E.; Glaunsinger, B.A. Infection-induced retrotransposon-derived noncoding RNAs enhance herpesviral gene expression via the NF-κB pathway. PLoS Pathog. 2015, 11, e1005260. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Chen, Z.J. Innate immune sensing and signaling of cytosolic nucleic acids. Annu. Rev. Immunol. 2014, 32, 461–488. [Google Scholar] [CrossRef] [PubMed]
- Dong, X.; Feng, H.; Sun, Q.; Li, H.; Wu, T.-T.; Sun, R.; Tibbetts, S.A.; Chen, Z.J.; Feng, P. Murine gamma-herpesvirus 68 hijacks MAVS and IKKβ to initiate lytic replication. PLoS Pathog. 2010, 6, e1001001. [Google Scholar] [CrossRef] [PubMed]
- Hung, T.; Pratt, G.A.; Sundararaman, B.; Townsend, M.J.; Chaivorapol, C.; Bhangale, T.; Graham, R.R.; Ortmann, W.; Criswell, L.A.; Yeo, G.W.; et al. The Ro60 autoantigen binds endogenous retroelements and regulates inflammatory gene expression. Science 2015, 350, 455–459. [Google Scholar] [CrossRef] [PubMed]
- Kaneko, H.; Dridi, S.; Tarallo, V.; Gelfand, B.D.; Fowler, B.J.; Cho, W.G.; Kleinman, M.E.; Ponicsan, S.L.; Hauswirth, W.W.; Chiodo, V.A.; et al. DICER1 deficit induces Alu RNA toxicity in age-related macular degeneration. Nature 2011, 471, 325–330. [Google Scholar] [CrossRef] [PubMed]
- Tarallo, V.; Hirano, Y.; Gelfand, B.D.; Dridi, S.; Kerur, N.; Kim, Y.; Cho, W.G.; Kaneko, H.; Fowler, B.J.; Bogdanovich, S.; et al. DICER1 loss and Alu RNA induce age-related macular degeneration via the NLRP3 inflammasome and MyD88. Cell 2012, 149, 847–859. [Google Scholar] [CrossRef] [PubMed]
- Karijolich, J.; Zhao, Y.; Alla, R.; Glaunsinger, B. Genome-wide mapping of infection-induced SINE RNAs reveals a role in selective mRNA export. Nucleic Acids Res. 2017, 45, 6194–6208. [Google Scholar] [CrossRef] [PubMed]
- Yeganeh, M.; Praz, V.; Cousin, P.; Hernandez, N. Transcriptional interference by RNA polymerase III affects expression of the Polr3e gene. Genes Dev. 2017, 31, 413–421. [Google Scholar] [CrossRef] [PubMed]
- Shearwin, K.E.; Callen, B.P.; Egan, J.B. Transcriptional interference—A crash course. Trends Genet. 2005, 21, 339–345. [Google Scholar] [CrossRef] [PubMed]
- Harreman, M.; Taschner, M.; Sigurdsson, S.; Anindya, R.; Reid, J.; Somesh, B.; Kong, S.E.; Banks, C.A.S.; Conaway, R.C.; Conaway, J.W.; et al. Distinct ubiquitin ligases act sequentially for RNA polymerase II polyubiquitylation. Proc. Natl. Acad. Sci. USA 2009, 106, 20705–20710. [Google Scholar] [CrossRef] [PubMed]
- Somesh, B.P.; Reid, J.; Liu, W.-F.; Søgaard, T.M.M.; Erdjument-Bromage, H.; Tempst, P.; Svejstrup, J.Q. Multiple mechanisms confining RNA polymerase II ubiquitylation to polymerases undergoing transcriptional arrest. Cell 2005, 121, 913–923. [Google Scholar] [CrossRef] [PubMed]
- Woudstra, E.C.; Gilbert, C.; Fellows, J.; Jansen, L.; Brouwer, J.; Erdjument-Bromage, H.; Tempst, P.; Svejstrup, J.Q. A Rad26-Def1 complex coordinates repair and RNA pol II proteolysis in response to DNA damage. Nature 2002, 415, 929–933. [Google Scholar] [CrossRef] [PubMed]
- Anindya, R.; Aygün, O.; Svejstrup, J.Q. Damage-induced ubiquitylation of human RNA polymerase II by the ubiquitin ligase Nedd4, but not Cockayne syndrome proteins or BRCA1. Mol. Cell 2007, 28, 386–397. [Google Scholar] [CrossRef] [PubMed]
- Jones, R.B.; Song, H.; Xu, Y.; Garrison, K.E.; Buzdin, A.A.; Anwar, N.; Hunter, D.V.; Mujib, S.; Mihajlovic, V.; Martin, E.; et al. LINE-1 retrotransposable element DNA accumulates in HIV-1-infected cells. J. Virol. 2013, 87, 13307–13320. [Google Scholar] [CrossRef] [PubMed]



© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dunker, W.; Zhao, Y.; Song, Y.; Karijolich, J. Recognizing the SINEs of Infection: Regulation of Retrotransposon Expression and Modulation of Host Cell Processes. Viruses 2017, 9, 386. https://doi.org/10.3390/v9120386
Dunker W, Zhao Y, Song Y, Karijolich J. Recognizing the SINEs of Infection: Regulation of Retrotransposon Expression and Modulation of Host Cell Processes. Viruses. 2017; 9(12):386. https://doi.org/10.3390/v9120386
Chicago/Turabian StyleDunker, William, Yang Zhao, Yu Song, and John Karijolich. 2017. "Recognizing the SINEs of Infection: Regulation of Retrotransposon Expression and Modulation of Host Cell Processes" Viruses 9, no. 12: 386. https://doi.org/10.3390/v9120386
APA StyleDunker, W., Zhao, Y., Song, Y., & Karijolich, J. (2017). Recognizing the SINEs of Infection: Regulation of Retrotransposon Expression and Modulation of Host Cell Processes. Viruses, 9(12), 386. https://doi.org/10.3390/v9120386