Estimation of Forest Canopy Height and Aboveground Biomass from Spaceborne LiDAR and Landsat Imageries in Maryland
Abstract
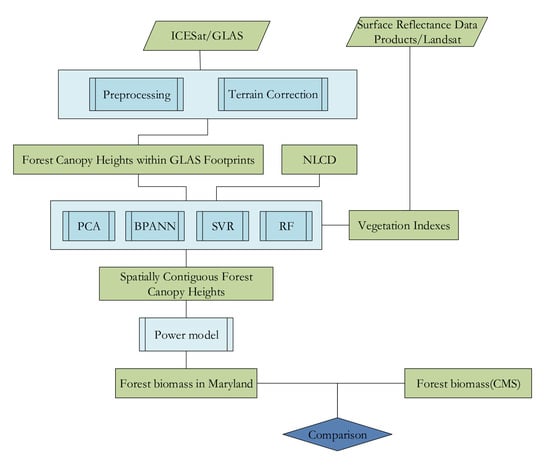
:1. Introduction
2. Study Area and Datasets
2.1. Study Area
2.2. ICESat GLAS Laser Altimetry Data
2.3. CMS_RF Forest Canopy Height and Aboveground Biomass for Maryland
2.4. Landsat Data
2.5. National Land Cover Database 2011
3. Methods
3.1. Estimating Forest Canopy Height from the GLAS Waveform
3.2. Extrapolating Canopy Height to the Study Area
3.3. Estimating Forest Aboveground Biomass in Maryland
4. Results
4.1. GLAS Waveform to Forest Canopy Height
4.2. Extrapolating Canopy Height to the Study Area
4.3. Forest Canopy Height to Aboveground Biomass
4.4. Comparing Biomass Estimates to Other Data Resources
5. Discussion
5.1. Processing the GLAS Waveform
5.2. Extrapolating to A Larger Spatial Scale with Spectral Images
5.3. Linking Forest Canopy Height to Biomass
5.4. Future Development
6. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
Appendix A
| Cross Validation | no.1 | no.2 | no.3 | no.4 | no.5 | no.6 | no.7 | no.8 | no.9 | no.10 | Average Value |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 5,t,l,tlm | 0.441 | 0.328 | 0.439 | 0.422 | 0.443 | 0.322 | 0.423 | 0.479 | 0.340 | 0.289 | 0.392 |
| 8,t,l,tlm | 0.427 | 0.420 | 0.453 | 0.399 | 0.438 | 0.406 | 0.396 | 0.411 | 0.341 | 0.299 | 0.399 |
| 20,t,l,tlm | 0.234 | 0.362 | 0.497 | 0.430 | 0.435 | 0.411 | 0.270 | 0.426 | 0.326 | 0.265 | 0.366 |
| 50,t,l,tlm | 0.308 | 0.246 | 0.563 | 0.239 | 0.240 | 0.377 | 0.207 | 0.330 | 0.238 | 0.251 | 0.300 |
| 8,t,t,tlm | 0.418 | 0.383 | 0.473 | 0.435 | 0.452 | 0.438 | 0.402 | 0.408 | 0.330 | 0.310 | 0.405 |
| 8,t,l,tlm | 0.461 | 0.409 | 0.454 | 0.446 | 0.409 | 0.414 | 0.385 | 0.451 | 0.286 | 0.337 | 0.405 |
| 8,l,t,tlm | 0.413 | 0.398 | 0.464 | 0.424 | 0.437 | 0.418 | 0.381 | 0.268 | 0.345 | 0.290 | 0.384 |
| 8,l,l,tlm | 0.468 | 0.368 | 0.457 | 0.402 | 0.458 | 0.419 | 0.376 | 0.393 | 0.328 | 0.307 | 0.398 |
| 8,l,p,tlm | 0.481 | 0.434 | 0.425 | 0.439 | 0.440 | 0.433 | 0.409 | 0.500 | 0.373 | 0.352 | 0.429 |
| 8,p,t,tlm | 0.461 | 0.403 | 0.430 | 0.438 | 0.442 | 0.436 | 0.383 | 0.485 | 0.339 | 0.326 | 0.414 |
| 8,p,l,tlm | 0.214 | 0.436 | 0.420 | 0.413 | 0.434 | 0.438 | 0.417 | 0.430 | 0.321 | 0.339 | 0.389 |
| 8,p,p,tlm | 0.468 | 0.413 | 0.391 | 0.401 | 0.403 | 0.414 | 0.418 | 0.457 | 0.350 | 0.328 | 0.404 |
| 8,t,l,tgd | 0.408 | 0.051 | 0.287 | 0.217 | 0.206 | 0.321 | 0.216 | 0.301 | 0.013 | 0.256 | 0.228 |
| 8,t,l,tgdx | 0.448 | 0.412 | 0.382 | 0.396 | 0.408 | 0.400 | 0.408 | 0.469 | 0.344 | 0.352 | 0.402 |
| 8,t,l,tgda | 0.462 | 0.407 | 0.372 | 0.373 | 0.370 | 0.424 | 0.397 | 0.423 | 0.339 | 0.306 | 0.387 |
| 8,t,l,trp | 0.477 | 0.426 | 0.422 | 0.445 | 0.432 | 0.436 | 0.421 | 0.481 | 0.357 | 0.345 | 0.424 |
| 8,t,l,tlm | 0.449 | 0.402 | 0.446 | 0.439 | 0.422 | 0.408 | 0.381 | 0.454 | 0.338 | 0.335 | 0.407 |
| 8,t,l,tfg | 0.463 | 0.429 | 0.435 | 0.455 | 0.436 | 0.442 | 0.423 | 0.470 | 0.375 | 0.351 | 0.428 |
| 8,t,l,tcg | 0.478 | 0.427 | 0.427 | 0.443 | 0.419 | 0.436 | 0.418 | 0.469 | 0.345 | 0.364 | 0.423 |
| 8,t,l,tss | 0.480 | 0.425 | 0.422 | 0.439 | 0.421 | 0.444 | 0.414 | 0.452 | 0.363 | 0.357 | 0.422 |
| 8,t,l,tgf | 0.470 | 0.365 | 0.425 | 0.347 | 0.427 | 0.443 | 0.417 | 0.478 | 0.352 | 0.352 | 0.408 |
| 8,t,l,tgp | 0.401 | 0.433 | 0.406 | 0.412 | 0.433 | 0.434 | 0.409 | 0.476 | 0.359 | 0.355 | 0.412 |
| Cross Validation | no.1 | no.2 | no.3 | no.4 | no.5 | no.6 | no.7 | no.8 | no.9 | no.10 | Average Value |
|---|---|---|---|---|---|---|---|---|---|---|---|
| -s 4 -t 2 -c 1 -g 1 -n 0.5 | 0.477 | 0.418 | 0.405 | 0.416 | 0.383 | 0.414 | 0.406 | 0.466 | 0.336 | 0.332 | 0.405 |
| -s 3 -t 0 -c 1 -g 1 -p 0.1 | 0.468 | 0.410 | 0.389 | 0.400 | 0.401 | 0.413 | 0.419 | 0.452 | 0.350 | 0.328 | 0.403 |
| -s 3 -t 1 -c 1 -g 1 -p 0.1 | 0.455 | 0.378 | 0.379 | 0.394 | 0.418 | 0.406 | 0.379 | 0.427 | 0.363 | 0.339 | 0.394 |
| -s 3 -t 2 -c 1 -g 1 -p 0.1 | 0.471 | 0.425 | 0.410 | 0.422 | 0.389 | 0.426 | 0.412 | 0.466 | 0.330 | 0.344 | 0.409 |
| Cross Validation | no.1 | no.2 | no.3 | no.4 | no.5 | no.6 | no.7 | no.8 | no.9 | no.10 | Average Value |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 10_2_1 | 0.407 | 0.376 | 0.655 | 0.432 | 0.416 | 0.402 | 0.389 | 0.435 | 0.409 | 0.396 | 0.432 |
| 50_2_1 | 0.480 | 0.410 | 0.689 | 0.459 | 0.439 | 0.418 | 0.393 | 0.468 | 0.456 | 0.409 | 0.462 |
| 100_2_1 | 0.482 | 0.419 | 0.700 | 0.469 | 0.442 | 0.417 | 0.398 | 0.471 | 0.452 | 0.420 | 0.467 |
| 200_2_1 | 0.478 | 0.424 | 0.704 | 0.475 | 0.442 | 0.424 | 0.395 | 0.476 | 0.449 | 0.418 | 0.468 |
| 500_2_1 | 0.478 | 0.423 | 0.707 | 0.473 | 0.446 | 0.428 | 0.396 | 0.482 | 0.445 | 0.423 | 0.470 |
| 1000_2_1 | 0.480 | 0.423 | 0.706 | 0.475 | 0.445 | 0.430 | 0.396 | 0.482 | 0.443 | 0.423 | 0.470 |
| 100_1_1 | 0.481 | 0.419 | 0.693 | 0.470 | 0.451 | 0.424 | 0.392 | 0.474 | 0.440 | 0.412 | 0.466 |
| 100_3_1 | 0.472 | 0.414 | 0.708 | 0.465 | 0.439 | 0.414 | 0.393 | 0.487 | 0.438 | 0.427 | 0.466 |
| 100_4_1 | 0.461 | 0.415 | 0.712 | 0.447 | 0.447 | 0.413 | 0.399 | 0.476 | 0.432 | 0.429 | 0.463 |
| 100_5_1 | 0.457 | 0.421 | 0.713 | 0.445 | 0.429 | 0.411 | 0.388 | 0.474 | 0.434 | 0.424 | 0.460 |
| 100_2_0 | 0.472 | 0.414 | 0.707 | 0.465 | 0.429 | 0.411 | 0.395 | 0.486 | 0.440 | 0.428 | 0.465 |
References
- Boudreau, J.; Nelson, R.F.; Margolis, H.A.; Beaudoin, A.; Guindon, L.; Kimes, D.S. Regional aboveground forest biomass using airborne and spaceborne LiDAR in Québec. Remote Sens. Environ. 2008, 112, 3876–3890. [Google Scholar] [CrossRef]
- Hamburg, S.P.; Zamolodchikov, D.G.; Korovin, G.N.; Nefedjev, V.V.; Utkin, A.I.; Gulbe, J.I.; Gulbe, T.A. Estimating the carbon content of Russian forests; a comparison of phytomass/volume and allometric projections. Mitig. Adapt. Strateg. Glob. Chang. 1997, 2, 247–265. [Google Scholar] [CrossRef]
- Ding, H.; Nunes, P.A.; Teelucksingh, S.S. European forests and carbon sequestration services: An economic assessment of climate change impacts. SSRN Electron. J. 2010. [Google Scholar] [CrossRef]
- Lasky, J.R.; Uriarte, M.; Boukili, V.K.; Erickson, D.L.; John Kress, W.; Chazdon, R.L. The relationship between tree biodiversity and biomass dynamics changes with tropical forest succession. Ecol. Lett. 2014, 17, 1158–1167. [Google Scholar] [CrossRef] [PubMed]
- Vance-Chalcraft, H.D.; Willig, M.R.; Cox, S.B.; Lugo, A.E.; Scatena, F.N. Relationship between aboveground biomass and multiple measures of biodiversity in subtropical forest of Puerto Rico. Biotropica 2010, 42, 290–299. [Google Scholar] [CrossRef]
- Nelson, R.; Parker, G.; Hom, M. A portable airborne laser system for forest inventory. Photogramm. Eng. Remote Sens. 2003, 69, 267–273. [Google Scholar] [CrossRef]
- Botkin, D.B.; Simpson, L.G. Biomass of the North American boreal forest: A step toward accurate global measures. Biogeochemistry 1990, 9, 161–174. [Google Scholar]
- Botkin, D.B.; Simpson, L.G.; Nisbet, R.A. Biomass and carbon storage of the North American deciduous forest. Biogeochemistry 1993, 20, 1–17. [Google Scholar] [CrossRef]
- Jenkins, J.C.; Chojnacky, D.C.; Heath, L.S.; Birdsey, R.A. National-scale biomass estimators for United States tree species. For. Sci. 2003, 49, 12–35. [Google Scholar]
- Penner, M.; Power, K.; Muhairwe, C.; Tellier, R.; Wang, Y. Canada’s Forest Biomass Resources: Deriving Estimates from Canada Forest Inventory; Information Report BC-X-370; Pacific Forestry Centre: Victoria, BC, Canada, 1997.
- Naesset, E. Airborne laser scanning as a method in operational forest inventory: Status of accuracy assessments accomplished in scandinavia. Scand. J. For. Res. 2007, 22, 433–442. [Google Scholar] [CrossRef]
- Næsset, E. Practical large-scale forest stand inventory using a small-footprint airborne scanning laser. Scand. J. For. Res. 2004, 19, 164–179. [Google Scholar] [CrossRef]
- Thomas, V.; Treitz, P.; Mccaughey, J.H.; Morrison, I. Mapping stand-level forest biophysical variables for a mixedwood boreal forest using lidar: An examination of scanning density. Can. J. For. Res. 2006, 36, 34–47. [Google Scholar] [CrossRef]
- Nelson, R. Measuring biomass and carbon in delaware using an airborne profiling lidar. Scand. J. For. Res. 2004, 19, 500–511. [Google Scholar] [CrossRef]
- Næsset, E.; Gobakken, T. Estimation of above- and below-ground biomass across regions of the boreal forest zone using airborne laser. Remote Sens. Environ. 2008, 112, 3079–3090. [Google Scholar] [CrossRef]
- Drake, J.B.; Dubayah, R.O.; Knox, R.G.; Clark, D.B.; Blair, J.B. Sensitivity of large-footprint lidar to canopy structure and biomass in a neotropical rainforest. Remote Sens. Environ. 2002, 81, 378–392. [Google Scholar] [CrossRef]
- Castro, K.L.; Sanchez-Azofeifa, G.A.; Rivard, B. Monitoring secondary tropical forests using space-borne data: Implications for Central America. Int. J. Remote Sens. 2003, 24, 1853–1894. [Google Scholar] [CrossRef]
- Harding, D.J.; Carabajal, C.C. ICESat waveform measurements of within-footprint topographic relief and vegetation vertical structure. Geophys. Res. Lett. 2005, 32, 741–746. [Google Scholar] [CrossRef]
- Brenner, A.; Zwally, H.; Bentley, C.; Csathó, B.; Harding, D.; Hofton, M.; Minster, J.; Roberts, L.; Saba, J.; Thomas, R.; et al. Derivation of Range and Range Distributions From Laser Pulse Waveform Analysis for Surface Elevations, Roughness, Slope, and Vegetation Heights. In Algorithm Theoretical Basis Document V4. 1; Goddard Space Flight Center: Greenbelt, MD, USA, 2003; p. 92. [Google Scholar]
- Fayad, I.; Baghdadi, N.; Bailly, J.S.; Barbier, N.; Gond, V.; Hérault, B.; El Hajj, M.; Lochard, J.; Perrin, J. Regional scale rain-forest height mapping using regression-kriging of spaceborne and airborne LiDAR data: Application on French Guiana. In Proceedings of the 2015 IEEE International Geoscience and Remote Sensing Symposium (IGARSS), Milan, Italy, 26–31 July 2015; pp. 4109–4112. [Google Scholar]
- Park, H.J.; Shin, H.S.; Roh, Y.H.; Kim, K.M.; Park, K.H. Estimating forest carbon stocks in Danyang using Kriging methods for aboveground biomass. J. Korean Assoc. Geogr. Inf. Stud. 2012, 15, 16–33. [Google Scholar] [CrossRef]
- Hajj, M.E.; Baghdadi, N.; Fayad, I.; Vieilledent, G.; Bailly, J.S.; Minh, D.H.T. Interest of integrating spaceborne LiDAR data to improve the estimation of biomass in high biomass forested areas. Remote Sens. 2017, 9, 213. [Google Scholar] [CrossRef]
- Chi, H.; Sun, G.; Huang, J.; Li, R.; Ren, X.; Ni, W.; Fu, A. Estimation of Forest Aboveground Biomass in Changbai Mountain Region Using ICESat/GLAS and Landsat/TM Data. Remote Sens. 2017, 9, 707. [Google Scholar] [CrossRef]
- Liu, K.; Wang, J.; Zeng, W.; Song, J. Comparison and Evaluation of Three Methods for Estimating Forest above Ground Biomass Using TM and GLAS Data. Remote Sens. 2017, 9, 341. [Google Scholar] [CrossRef]
- Dhanda, P.; Nandy, S.; Kushwaha, S.P.S.; Ghosh, S.; Murthy, Y.K.; Dadhwal, V.K. Optimizing spaceborne LiDAR and very high resolution optical sensor parameters for biomass estimation at ICESat/GLAS footprint level using regression algorithms. Prog. Phys. Geogr. 2017, 2, 1–21. [Google Scholar] [CrossRef]
- Nelson, R.; Margolis, H.; Montesano, P.; Sun, G.; Cook, B.; Corp, L.; Andersen, H.; Jong, B.; Pellat, F.P.; Fickl, T.; et al. Lidar-based estimates of aboveground biomass in the continental US and Mexico using ground, airborne, and satellite observations. Remote Sens. Environ. 2017, 188, 127–140. [Google Scholar] [CrossRef]
- Helmer, E.H.; Lefsky, M.A.; Roberts, D.A. Biomass accumulation rates of amazonian secondary forest and biomass of old-growth forests from landsat time series and the geoscience laser altimeter system. J. Appl. Remote Sens. 2009, 3, 201–210. [Google Scholar]
- Zhang, Y.; Liang, S.; Sun, G. Forest biomass mapping of northeastern China using GLAS and MODIS data. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2014, 7, 140–152. [Google Scholar] [CrossRef]
- Fu, A.; Sun, G.; Guo, Z. Estimating forest biomass with GLAS samples and MODIS imagery in northeastern China. Proc. SPIE 2009, 7498, 1–8. [Google Scholar]
- Huang, K.; Pang, Y.; Shu, Q.; Fu, T. Aboveground forest biomass estimation using ICESat GLAS in Yunnan, China. Yaogan Xuebao-J. Remote Sens. 2013, 17, 165–179. [Google Scholar]
- Yavaşlı, D.D. Estimation of above ground forest biomass at Muğla using ICESat/GLAS and Landsat data. Remote Sens. Appl. Soc. Environ. 2016, 4, 211–218. [Google Scholar] [CrossRef]
- Hermosilla, T.; Wulder, M.A.; White, J.C.; Coops, N.C.; Hobart, G.W. An integrated Landsat time series protocol for change detection and generation of annual gap-free surface reflectance composites. Remote Sens. Environ. 2015, 158, 220–234. [Google Scholar] [CrossRef]
- Armston, J.D.; Denham, R.J.; Danaher, T.J.; Scarth, P.F.; Moffiet, T.N. Prediction and validation of foliage projective cover from Landsat-5 TM and Landsat-7 ETM+ imagery. J. Appl. Remote Sens. 2009, 3, 033540. [Google Scholar] [CrossRef]
- Yan, L.; Roy, D.P. Conterminous United States crop field size quantification from multi-temporal Landsat data. Remote Sens. Environ. 2016, 172, 67–86. [Google Scholar] [CrossRef]
- Sesnie, S.E.; Gessler, P.E.; Finegan, B.; Thessler, S. Integrating Landsat TM and SRTM-DEM derived variables with decision trees for habitat classification and change detection in complex neotropical environments. Remote Sens. Environ. 2008, 112, 2145–2159. [Google Scholar] [CrossRef]
- Oyama, Y.; Matsushita, B.; Fukushima, T. Distinguishing surface cyanobacterial blooms and aquatic macrophytes using Landsat/TM and ETM+ shortwave infrared bands. Remote Sens. Environ. 2015, 157, 35–47. [Google Scholar] [CrossRef]
- Kovalskyy, V.; Roy, D.P. The global availability of Landsat 5 TM and Landsat 7 ETM+ land surface observations and implications for global 30 m Landsat data product generation. Remote Sens. Environ. 2013, 130, 280–293. [Google Scholar] [CrossRef]
- Feng, M.; Sexton, J.O.; Huang, C.; Masek, J.G.; Vermote, E.F.; Gao, F.; Narasimhan, R.; Channan, S.; Wolfe, R.E.; Townshend, J.R. Global surface reflectance products from Landsat: Assessment using coincident MODIS observations. Remote Sens. Environ. 2013, 134, 276–293. [Google Scholar] [CrossRef]
- Huang, W.; Swatantran, A.; Johnson, K.; Duncanson, L.; Tang, H.; Dunne, J.O.N.; Hurtt, G.; Dubayah, R. Local discrepancies in continental scale biomass maps: A case study over forested and non-forested landscapes in Maryland, USA. Carbon Balance Manag. 2015, 10, 19. [Google Scholar] [CrossRef] [PubMed]
- Weber, T.; Sloan, A.; Wolf, J. Maryland’s Green Infrastructure Assessment: Development of a comprehensive approach to land conservation. Landsc. Urban Plan. 2006, 77, 94–110. [Google Scholar] [CrossRef]
- O’Loughlin, F.E.; Paiva, R.C.D.; Durand, M.; Alsdorf, D.E.; Bates, P.D. A multi-sensor approach towards a global vegetation corrected SRTM DEM product. Remote Sens. Environ. 2016, 182, 49–59. [Google Scholar] [CrossRef]
- Schutz, B.E. Spaceborne laser altimetry: 2001 and beyond. In Book of Extended Abstracts WEGENER-98; Plag, H.P., Ed.; Norwegian Mapping Authority: Honefoss, Norway, 1998. [Google Scholar]
- Bye, I.J.; North, P.R.J.; Los, S.O.; Kljun, N.; Rosette, J.A.B.; Hopkinson, C.; Chasmer, L.; Mahoney, C. Estimating forest canopy parameters from satellite waveform LiDAR by inversion of the FLIGHT three-dimensional radiative transfer model. Remote Sens. Environ. 2017, 188, 177–189. [Google Scholar] [CrossRef]
- Schutz, B.E.; Zwally, H.J.; Shuman, C.A.; Hancock, D.; DiMarzio, J.P. Overview of the ICESat mission. Geophys. Res. Lett. 2005, 32, L21S01. [Google Scholar] [CrossRef]
- Chi, H.; Sun, G.; Huang, J.; Guo, Z.; Ni, W.; Fu, A. National forest aboveground biomass mapping from ICESat/GLAS data and MODIS imagery in China. Remote Sens. 2015, 7, 5534–5564. [Google Scholar] [CrossRef]
- Su, Y.; Guo, Q.; Xue, B.; Hu, T.; Alvarez, O.; Tao, S.; Fang, J. Spatial distribution of forest aboveground biomass in China: Estimation through combination of spaceborne lidar, optical imagery, and forest inventory data. Remote Sens. Environ. 2016, 173, 187–199. [Google Scholar] [CrossRef]
- Sun, G.; Ranson, K.J.; Kimes, D.S.; Blair, J.B.; Kovacs, K. Forest vertical structure from GLAS: An evaluation using LVIS and SRTM data. Remote Sens. Environ. 2008, 112, 107–117. [Google Scholar] [CrossRef]
- Neuenschwander, A.L.; Urban, T.J.; Gutierrez, R.; Schutz, B.E. Characterization of ICESat/GLAS waveforms over terrestrial ecosystems: Implications for vegetation mapping. J. Geophys. Res. Biogeosci. 2008, 113, 1032. [Google Scholar] [CrossRef]
- Hilbert, C.; Schmullius, C. Influence of surface topography on ICESat/GLAS forest height estimation and waveform shape. Remote Sens. 2012, 4, 2210–2235. [Google Scholar] [CrossRef]
- Lefsky, M.A.; Harding, D.J.; Keller, M.; Cohen, W.B.; Carabajal, C.C.; Del Bom Espirito-Santo, F.; Hunter, M.O.; de Oliveira, R., Jr.; de Camargo, P.B. Estimates of forest canopy height and aboveground biomass using ICESat. Geophys. Res. Lett. 2006, 33, L05501. [Google Scholar] [CrossRef]
- Fayad, I.; Baghdadi, N.; Gond, V.; Bailly, J.S.; Barbier, N.; El Hajj, M.; Fabre, F. Coupling potential of ICESat/GLAS and SRTM for the discrimination of forest landscape types in French Guiana. Int. J. Appl. Earth Obs. Geoinf. 2014, 33, 21–31. [Google Scholar] [CrossRef] [Green Version]
- Mahoney, C.; Kljun, N.; Los, S.O.; Chasmer, L.; Hacker, J.M.; Hopkinson, C.; North, P.; Rosette, J.; van Gorsel, E. Slope estimation from ICESat/GLAS. Remote Sens. 2014, 6, 10051–10069. [Google Scholar] [CrossRef]
- Dubayah, R.O.; Swatantran, A.; Huang, W.; Duncanson, L.; Johnson, K.; Tang, H.; Dunne, J.O.; Hurtt, G.C. CMS: LiDAR-Derived Aboveground Biomass, Canopy Height and Cover for Maryland, 2011; ORNL DAAC: Oak Ridge, TN, USA, 2016.
- O’Neil-Dunne, J.P.; MacFaden, S.W.; Royar, A.R.; Pelletier, K.C. An object-based system for LiDAR data fusion and feature extraction. Geocarto Int. 2014, 28, 227–242. [Google Scholar] [CrossRef]
- Dubayah, R. County-Scale Carbon Estimation in NASA’s Carbon Monitoring System. Biomass Carbon Storage 2012. Available online: https://www.researchgate.net/profile/Ross_Nelson/publication/258459476_County-Scale_Carbon_Estimation_in_NASA's_Carbon_Monitoring_System/links/564e2fe508ae1ef9296_c6779.pdf (accessed on 1 November 2017).
- Yan, L.; Roy, D.P. Automated crop field extraction from multi-temporal Web Enabled Landsat Data. Remote Sens. Environ. 2014, 144, 42–64. [Google Scholar] [CrossRef]
- Baumann, M.; Ozdogan, M.; Kuemmerle, T.; Wendland, K.J.; Esipova, E.; Radeloff, V.C. Using the Landsat record to detect forest-cover changes during and after the collapse of the Soviet Union in the temperate zone of European Russia. Remote Sens. Environ. 2012, 124, 174–184. [Google Scholar] [CrossRef]
- Melaas, E.K.; Friedl, M.A.; Zhu, Z. Detecting interannual variation in deciduous broadleaf forest phenology using Landsat TM/ETM + data. Remote Sens. Environ. 2013, 132, 176–185. [Google Scholar] [CrossRef]
- Lasanta, T.; Vicente-Serrano, S.M. Complex land cover change processes in semiarid Mediterranean regions: An approach using Landsat images in northeast Spain. Remote Sens. Environ. 2012, 124, 1–14. [Google Scholar] [CrossRef]
- Solomon, D.S.; Hosmer, R.A.; Hayslett, H.T., Jr. A two-stage matrix model for predicting growth of forest stands in the Northeast. Can. J. For. Res. 1986, 16, 521–528. [Google Scholar] [CrossRef]
- Clewley, D.; Lucas, R.; Accad, A.; Armston, J.; Bowen, M.; Dwyer, J.; Pollock, S.; Bunting, P.; McAlpine, C.; Kelly, A.; et al. An approach to mapping forest growth stages in Queensland, Australia through integration of ALOS PALSAR and Landsat sensor data. Remote Sens. 2012, 4, 2236–2255. [Google Scholar] [CrossRef] [Green Version]
- Powell, S.L.; Cohen, W.B.; Healey, S.P.; Kennedy, R.E.; Moisen, G.G.; Pierce, K.B.; Ohmann, J.L. Quantification of live aboveground forest biomass dynamics with Landsat time-series and field inventory data: A comparison of empirical modeling approaches. Remote Sens. Environ. 2010, 114, 1053–1068. [Google Scholar] [CrossRef]
- Wickham, J.; Stehman, S.V.; Gass, L.; Dewitz, J.A.; Sorenson, D.G.; Granneman, B.J.; Poss, R.V.; Baer, L.A. Thematic accuracy assessment of the 2011 national land cover database (NLCD). Remote Sens. Environ. 2017, 191, 328–341. [Google Scholar] [CrossRef]
- Xian, G.; Homer, C.; Dewitz, J.; Fry, J.; Hossain, N.; Wickham, J. Change of impervious surface area between 2001 and 2006 in the conterminous United States. Photogramm. Eng. Remote Sens. 2011, 77, 758–762. [Google Scholar]
- Sun, G.; Masek, J.; Guo, Z.; Pang, Y.; Fu, A.; Wang, D. Estimation of tree height and forest biomass from GLAS data. J. For. Plan. 2008, 13, 157–164. [Google Scholar]
- Chen, Q. Retrieving vegetation height of forests and woodlands over mountainous areas in the pacific coast region using satellite laser altimetry. Remote Sens. Environ. 2010, 114, 1610–1627. [Google Scholar] [CrossRef]
- Valor, E.; Caselles, V. Mapping land surface emissivity from NDVI: Application to European, African, and South American areas. Remote Sens. Environ. 1996, 57, 167–184. [Google Scholar] [CrossRef]
- Matsushita, B.; Yang, W.; Chen, J.; Onda, Y.; Qiu, G. Sensitivity of the enhanced vegetation index (EVI) and normalized difference vegetation index (NDVI) to topographic effects: A case study in high-density cypress forest. Sensors 2007, 7, 2636–2651. [Google Scholar] [CrossRef] [PubMed]
- Tucker, C.J. Red and photographic infrared linear combinations for monitoring vegetation. Remote Sens. Environ. 1979, 8, 127–150. [Google Scholar] [CrossRef]
- Gupta, R.K. Comparative study of AVHRR ratio vegetation index and normalized difference vegetation index in district level agricultural monitoring. Int. J. Remote Sens. 1993, 14, 53–73. [Google Scholar] [CrossRef]
- Kaiser, J. MODIS-derived visible atmospherically resistant index for monitoring chaparral moisture content. Int. J. Remote Sens. 2005, 26, 3867–3873. [Google Scholar]
- Hammer, O.; Harper, D.A.; Ryan, P.D. Past: Paleontological statistics software package for education and data analysis. Palaeontol. Electron. 2001, 4, 1–9. [Google Scholar]
- Roweis, S. EM algorithms for PCA and SPCA. Adv. Neural Inf. Process. Syst. 1998, 10, 626–632. [Google Scholar]
- Mika, S.; Schölkopf, B.; Smola, A.J.; Müller, K.R.; Scholz, M.; Rätsch, G. Kernel PCA and de-noising in feature spaces. In Advances in Neural Information Processing Systems; The MIT Press: Cambridge, MA, USA, 1999; pp. 536–542. [Google Scholar]
- Rumelhart, D.E.; Hinton, G.E.; Williams, R.J. Learning representations by back-propagating errors. Nature 1986, 323, 533–538. [Google Scholar] [CrossRef]
- Sun, G.; Hoff, S.J.; Zelle, B.C.; Smith, M.A. Development and comparison of backpropagation and generalized regression neural network models to predict diurnal and seasonal gas and PM10 concentrations and emissions from swine buildings. Trans. Asabe 2008, 51, 685–694. [Google Scholar] [CrossRef]
- Tamilarasan, A.; Mukkamala, S.; Sung, A.H.; Yendrapalli, K. Feature ranking and selection for intrusion detection using artificial neural networks and statistical methods. In Proceedings of the International Joint Conference on Neural Networks, Vancouver, BC, Canada, 16–21 July 2006; pp. 4754–4761. [Google Scholar]
- Chen, P.H.; Lin, C.J.; Schölkopf, B. A tutorial on ν-support vector machines. Appl. Stoch. Models Bus. Ind. 2005, 21, 111–136. [Google Scholar] [CrossRef]
- Campbell, W.M.; Sturim, D.E.; Reynolds, D.A.; Solomonoff, A. SVM Based Speaker Verification using a GMM Supervector Kernel and NAP Variability Compensation. In Proceedings of the IEEE International Conference on Acoustics, Speech, and Signal Processing, Toulouse, France, 14–19 May 2006; Volume 1, pp. 97–100. [Google Scholar]
- Chang, C.C.; Lin, C.J. Training nu-support vector regression: Theory and algorithms. Neural Comput. 2002, 14, 1959–1977. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Song, A.; Xiao, J. A robust static decoupling algorithm for 3-axis force sensors based on coupling error model and ε-SVR. Sensors 2012, 12, 14537–14555. [Google Scholar] [CrossRef] [PubMed]
- Breiman, L. Random forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Jiang, P.; Wu, H.; Wei, J.; Sang, F.; Sun, X.; Lu, Z. RF-DYMHC: Detecting the yeast meiotic recombination hotspots and coldspots by random forest model using gapped dinucleotide composition features. Nucleic Acids Res. 2007, 35, W47–W51. [Google Scholar] [CrossRef] [PubMed]
- Saatchi, S.S.; Harris, N.L.; Brown, S.; Lefsky, M.; Mitchard, E.T.; Salas, W. Benchmark map of forest carbon stocks in tropical regions across three continents. Proc. Natl. Acad. Sci. USA 2011, 108, 9899–9904. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Ni-Meister, W.; Yang, W.; Chen, Q. Physically based vertical vegetation structure retrieval from ICESat data: Validation using LVIS in White Mountain National Forest, New Hampshire, USA. Remote Sens. Environ. 2011, 115, 2776–2785. [Google Scholar] [CrossRef]
- Cao, C.; Ni, X.; Wang, X.; Lu, S.; Zhang, Y.; Dang, Y.; Singh, R.P. Allometric scaling theory-based maximum forest tree height and biomass estimation in the Three Gorges reservoir region using multi-source remote-sensing data. Int. J. Remote Sens. 2016, 37, 1210–1222. [Google Scholar] [CrossRef]














© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, M.; Sun, R.; Xiao, Z. Estimation of Forest Canopy Height and Aboveground Biomass from Spaceborne LiDAR and Landsat Imageries in Maryland. Remote Sens. 2018, 10, 344. https://doi.org/10.3390/rs10020344
Wang M, Sun R, Xiao Z. Estimation of Forest Canopy Height and Aboveground Biomass from Spaceborne LiDAR and Landsat Imageries in Maryland. Remote Sensing. 2018; 10(2):344. https://doi.org/10.3390/rs10020344
Chicago/Turabian StyleWang, Mengjia, Rui Sun, and Zhiqiang Xiao. 2018. "Estimation of Forest Canopy Height and Aboveground Biomass from Spaceborne LiDAR and Landsat Imageries in Maryland" Remote Sensing 10, no. 2: 344. https://doi.org/10.3390/rs10020344
APA StyleWang, M., Sun, R., & Xiao, Z. (2018). Estimation of Forest Canopy Height and Aboveground Biomass from Spaceborne LiDAR and Landsat Imageries in Maryland. Remote Sensing, 10(2), 344. https://doi.org/10.3390/rs10020344