Sentinel-2 Remote Sensed Image Classification with Patchwise Trained ConvNets for Grassland Habitat Discrimination
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Area and Grassland Habitats Characterization
2.2. Data Availability
2.2.1. Ground Truth
2.2.2. Satellite Data
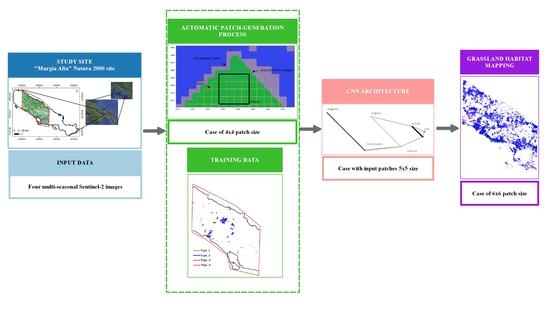
2.3. Algorithm for Habitat Mapping
2.3.1. CNN Classifier Configuration
- The input layer specifies the size of the patches, that in our case is variable between 1 × 1 and 6 × 6, while num_bands (in our case equal to 40) refers the depth of the multispectral Sentinel data;
- kernel_size is an integer specifying the height and width of the 2D convolution window, whereas depth (equal to 32 in our case) is the dimensionality of the output space (i.e., the number of output filters in the convolution). Such value was determined in the meta-parameter tuning procedure as an effective compromise between the architecture complexity and the learning curve convergence;
- output_size (equal to 128 in our case) is the number of output neurons of the first FC layer;
- num_classes (equal to 4 in our case) is the number of output neurons of the latest FC layer.
2.3.2. Experimental Setting
- A.
- Evaluating how the information needed for classification spreads over multiple multi-spectral pixels varying the size of square input patches to the ConvNet (Information Localization). In detail, in our setting the kernel size of the ConvNet (kernel_size × kernel_size) was grown linearly with the patch size. As no padding was set up, the FC part of the network remained unchanged while the convolution kernel increased and took charge of the pattern recognition task.
- B.
- Comparing the performance of a ConvNet with that of a corresponding FC architecture network. For fair comparison, the FC was set up by leaving untouched the original ConvNet with the exception of the kernel size of the convolutional layer, which, in this second instance, has been kept to a 1 × 1 size.Our CNN settings include:
- A total of 120 epochs with a batch size of 32 and 1000 steps per epoch;
- A kernel size equal to the size of the input patches;
- An Adadelta optimizer with: (a) 0.001 as learning rate; (b) a decay rate of 0.95; (c) a stability factor of 1 × 10−7.
2.3.3. Accuracy Assessment
- TP = True Positive (number of samples correctly assigned to a class);
- FP = False Positive (number of samples incorrectly assigned to a class);
- FN = False Negative (number of samples of a class incorrectly assigned to another);
- The F1-score metric performs the harmonic mean of Precision and Recall [72].
3. Results
3.1. Grassland Habitats Characterization
3.2. Information Localization
3.3. ConvNet vs. FC Network Performance
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lu, D.; Weng, Q. A survey of image classification methods and techniques for improving classification performance. Int. J. Remote Sens. 2007, 28, 823–870. [Google Scholar] [CrossRef]
- Schuster, C.; Schmidt, T.; Conrad, C.; Kleinschmit, B.; Forster, M. Grassland habitat mapping by intra-annual time series analysis–Comparison of RapidEye and TerraSAR-X satellite data. Int. J. Appl. Earth Obs. Geoinf. 2015, 34, 25–34. [Google Scholar] [CrossRef]
- Ali, I.; Cawkwell, F.; Dwyer, E.; Barrett, B.; Green, S. Satellite remote sensing of grasslands: From observation to management. J. Plant Ecol. 2016, 9, 649–671. [Google Scholar] [CrossRef] [Green Version]
- Franke, J.; Keuck, V.; Siegert, F. Assessment of grassland use intensity by remote sensing to support conservation schemes. J. Nat. Conserv. 2012, 20, 125–134. [Google Scholar] [CrossRef]
- Dusseux, P.; Corpetti, T.; Hubet-Moy, L.; Corgne, S. Combined use of Multi-temporal optical and radar satellite images for grassland monitoring. Remote Sens. 2014, 6, 6163–6182. [Google Scholar] [CrossRef] [Green Version]
- Xu, D.; Chen, B.; Shen, B.; Wang, X.; Yan, Y.; Xu, L.; Xin, X. The Classification of Grassland Types Based on Object-Based Image Analysis with Multisource Data. Rangel. Ecol. Manag. 2019, 72, 318–326. [Google Scholar] [CrossRef]
- Melville, B.; Lucieer, A.; Aryal, J. Object-based random forest classification of Landsat ETM+ and worldview-2 satellite imagery for mapping lowland native grassland communities in Tasmania, Australia. Int. J. Appl. Earth Obs. Geoinf. 2018, 66, 46–55. [Google Scholar] [CrossRef]
- Buck, O.; Garcia Millàn, V.E.; Klink, A.; Pakzad, K. Using information layers for mapping grassland habitat distribution at local to regional scales. Int. J. Appl. Earth Obs. Geoinf. 2015, 37, 83–89. [Google Scholar] [CrossRef]
- Zlinszky, A.; Schroiff, A.; Kania, A.; Deák, B.; Mücke, W.; Vári, A.; Székely, B.; Pfeifer, N. Categorizing Grassland Vegetation with Full-Waveform Airborne Laser Scanning: A Feasibility Study for Detecting Natura 2000 Habitat Types. Remote Sens. 2014, 6, 8056–8087. [Google Scholar] [CrossRef] [Green Version]
- Marcinkowska-Ochtyra, A.; Gryguc, K.; Ochtyra, A.; Kopeć, D.; Jarocinska, A.; Sławik, L. Multitemporal Hyperspectral Data Fusion with Topographic Indices—Improving Classification of Natura 2000 Grassland Habitats. Remote Sens. 2019, 11, 2264. [Google Scholar] [CrossRef] [Green Version]
- Rapinel, S.; Mony, C.; Lecoq, L.; Clément, B.; Thomas, A.; Hubert-Moy, L. Evaluation of Sentinel-2 time-series for mapping floodplain grassland plant communities. Remote Sens. Environ. 2019, 223, 115–129. [Google Scholar] [CrossRef]
- Fauvel, M.; Lopes, M.; Dubo, T.; Rivers-Moore, J.; Frison, P.; Gross, N.; Ouin, A. Prediction of plant diversity in grasslands using Sentinel-1 and -2 satellite image time-series. Remote Sens. Environ. 2020, 237, 111536. [Google Scholar] [CrossRef]
- Tarantino, C.; Forte, L.; Blonda, P.; Vicario, S.; Tomaselli, V.; Beierkuhnlein, C.; Adamo, M. Intra-Annual Sentinel-2 Time-Series Supporting Grassland Habitat Discrimination. Remote Sens. 2021, 13, 277. [Google Scholar] [CrossRef]
- Dixon, B.; Candade, N. Multispectral land use classification using neural networks and support vector machines: One or the other, or both? Int. J. Rem. Sens. 2008, 29, 1185–1206. [Google Scholar] [CrossRef]
- Kanellopoulos, I.; Wilkinson, G.G. Strategies and best practice for neural network image classification. Int. J. Remote Sens. 2010, 18, 711–725. [Google Scholar] [CrossRef]
- Jarvis, C.H.; Stuart, N. The sensitivity of a neural network for classifying remotely sensed imagery. Comput. Geosci. 1996, 22, 959–967. [Google Scholar] [CrossRef]
- Zhou, L.; Yang, X. An Assessment of Internal Neural Network Parameters Affecting Image Classification Accuracy. Remote Sens. 2011, 77, 12. [Google Scholar] [CrossRef]
- Chen, X.-W.; Lin, X. Big Data Deep Learning: Challenges and Perspectives. IEEE Access 2014, 2, 514–525. [Google Scholar] [CrossRef]
- Lecun, Y.; Bottou, L.; Bengio, Y.; Haffner, P. Gradient-based learning applied to document recognition. Proc. IEEE 1998, 86, 2278–2324. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.; Zhang, Y. Classification of hyperspectral image based on deep belief networks. In Proceedings of the 2014 IEEE International Conference on Image Processing (ICIP), Paris, France, 27–30 October 2014. [Google Scholar] [CrossRef]
- Hu, F.; Xia, G.-S.; Hu, J.; Zhang, L. Transferring Deep Convolutional Neural Networks for the Scene Classification of High-Resolution Remote Sensing Imagery. Remote Sens. 2015, 7, 14680–14707. [Google Scholar] [CrossRef] [Green Version]
- Luus, F.P.S.; Salmon, B.P.; van den Bergh, F.; Maharaj, B.T.J. Multiview Deep Learning for Land-Use Classification. IEEE Geosci. Remote Sens. Lett. 2015, 12, 2448–2452. [Google Scholar] [CrossRef] [Green Version]
- Ruiz Emparanza, P.; Hongkarnjanakul, N.; Rouquette, D.; Schwob, C.; Mezeix, L. Land cover classification in Thailand’s Eastern Economic Corridor (EEC) using convolutional neural network on satellite images. Remote Sens. Appl. Soc. Environ. 2020, 20, 100394. [Google Scholar] [CrossRef]
- Pan, S.; Guan, H.; Chen, Y.; Yu, Y.; Gonçalves, W.N.; Junior, J.M.; Li, J. Land-cover classification of multispectral LiDAR data using CNN with optimized hyper-parameters. ISPRS J. Photogramm. Remote Sens. 2020, 166, 241–254. [Google Scholar] [CrossRef]
- Zhang, C.; Yue, P.; Tapete, D.; Shangguan, B.; Wang, M.; Wu, Z. A multi-level context-guided classification method with object-based convolutional neural network for land cover classification using very high resolution remote sensing images. Int. J. Appl. Earth Obs. Geoinf. 2020, 88, 102086. [Google Scholar] [CrossRef]
- Ma, L.; Liu, Y.; Zhang, X.; Ye, Y.; Yin, G.; Johnson, B.A. Deep learning in remote sensing applications: A meta-analysis and review. ISPRS J. Photogramm. Remote Sens. 2019, 152, 166–177. [Google Scholar] [CrossRef]
- Yoo, C.; Han, D.; Im, J.; Bechtel, B. Comparison between convolutional neural networks and random forest for local climate zone classification in mega urban areas using Landsat images. ISPRS J. Photogramm. Remote Sens. 2019, 157, 155–170. [Google Scholar] [CrossRef]
- Watanabe, F.S.; Miyoshi, G.T.; Rodrigues, T.W.; Bernardo, N.M.; Rotta, L.H.; Alcântara, E.; Imai, N.N. Inland water’s trophic status classification based on machine learning and remote sensing data. Remote Sens. Appl. Soc. Environ. 2020, 19, 100326. [Google Scholar] [CrossRef]
- Mairota, P.; Leronni, V.; Xi, W.; Mladenoff, D.; Nagendra, H. Using spatial simulations of habitat modification for adaptive management of protected areas: Mediterranean grassland modification by woody plant encroachment. Environ. Conserv. 2013, 41, 144–146. [Google Scholar] [CrossRef]
- Forte, L.; Perrino, E.V.; Terzi, M. Le praterie a Stipa austroitalica Martinovsky ssp. austroitalica dell’Alta Murgia (Puglia) e della Murgia Materana (Basilicata). Fitosociologia 2005, 42, 83–103. [Google Scholar]
- Council Directive 2009/147/EEC. Available online: https://eur-lex.europa.eu/legal-content/EN/TXT/?uri=CELEX%3A32009L0147 (accessed on 26 June 2019).
- Sutter, G.C.; Brigham, R.M. Avifaunal and habitat changes resulting from conversion of native prairie to crested wheat grass:Patterns at songbird community and species levels. Can. J. Zool. 1998, 76, 869–875. [Google Scholar] [CrossRef]
- Brotons, L.; Pons, P.; Herrando, S. Colonization of dynamic Mediterranean landscapes: Where do birds come from after fire? J. Biogeogr. 2005, 32, 789–798. [Google Scholar] [CrossRef]
- Mairota, P.; Cafarelli, B.; Labadessa, R.; Lovergine, F.; Tarantino, C.; Lucas, R.M.; Nagendra, H.; Didham, R.K. Very high resolution Earth observation features for monitoring plant and animal community structure across multiple spatial scales in protected areas. Int. J. Appl. Earth Obs. Geoinf. 2015, 37, 100–105. [Google Scholar] [CrossRef]
- Tarantino, C.; Casella, F.; Adamo, M.; Lucas, R.; Beierkuhnlein, C.; Blonda, P. Ailanthus altissima mapping from multi-temporal very high resolution satellite images. ISPRS J. Photogram. Remote Sens. 2019, 147, 90–103. [Google Scholar] [CrossRef]
- Council Directive 92/43/EEC. Available online: https://eur-lex.europa.eu/legal-content/EN/TXT/?uri=celex%3A31992L0043 (accessed on 1 July 2013).
- Davies, C.E.; Moss, D. EUNIS Habitat Classification. In Final Report to the European Topic Centre of Nature Protection and Biodiversity; European Environment Agency: Swindon, UK, 2002. [Google Scholar]
- Braun-Blanquet, J. Pflanzensoziologie: Grundzüge der Vegetationskunde: Plant Sociology Basics of Vegetation Science; Springer: Berlin/Heidelberg, Germany, 1964; pp. 287–399. [Google Scholar]
- EU. Habitats Manual, Interpretation Manual of European Union Habitats: 1–144. 2013. Available online: http://ec.europa.eu/environment/nature/legislation/habitatsdirective/docs/Int_Manual_EU28.pdf (accessed on 1 April 2013).
- Biondi, E.; Blasi, C.; Burrascano, S.; Casavecchia, S.; Copiz, R.; Del Vico, E.; Galdenzi, D.; Gigante, D.; Lasen, C.; Spampinato, G.; et al. Manuale Italiano di Interpretazione Degli Habitat Della Direttiva 92/43/CEE. MATTM-DPN, SBI. 2010. Available online: http://vnr.unipg.it/habitat/index.jsp (accessed on 1 December 2007).
- Westhoff, V.; van der Maarel, E. The Braun-Blanquet Approach. In Classification of Plant Communities; Whittaker, R.H., Ed.; Junk: The Hague, The Netherlands, 1978; pp. 287–399. [Google Scholar]
- Biondi, E.; Burrascano, S.; Casavecchia, S.; Copiz, R.; Del Vico, E.; Galdenzi, D.; Gigante, D.; Lasen, C.; Spampinato, G.; Venanzoni, R.; et al. Diagnosis and syntaxonomic interpretation of Annex I Habitats (Dir. 92/43/ EEC) in Italy at the alliance level. Plant Sociol. 2012, 49, 5–37. [Google Scholar]
- Biondi, E.; Blasi, C. Prodromo della Vegetazione Italiana 2015. Ministero dell’Ambiente e della Tutela del Territorio e del Mare. Available online: http://www.prodromo-vegetazione-italia.org/ (accessed on 1 March 2015).
- USGS Portal. Available online: https://earthexplorer.usgs.gov/ (accessed on 9 May 2018).
- Anagnostis, A.; Asiminari, G.; Papageorgiou, E.; Bochtis, D. A Convolutional Neural Networks Based Method for Anthracnose Infected Walnut Tree Leaves Identification. NATO Adv. Sci. Inst. Ser. E Appl. Sci. 2020, 10, 469. [Google Scholar] [CrossRef] [Green Version]
- Hasan, M.; Ullah, S.; Khan, M.J.; Khurshid, K. Comparative analysis of svm, ann and cnn for classifying vegetation species using hyperspectral thermal infrared data. In Proceedings of the ISPRS Geospatial Week 2019 (Volume XLII-2/W13), Enschede, The Netherlands, 10–14 June 2019; Copernicus GmbH: Göttingen, Germany, 2019; pp. 1861–1868. [Google Scholar]
- Rumelhart, D.E.; Hinton, G.E.; Williams, R.J. Learning Internal Representations by Error Propagation; Technical Report; California University San Diego; Institute for Cognitive Science: La Jolla, CA, USA, 1985; Available online: https://apps.dtic.mil/sti/pdfs/ADA164453.pdf (accessed on 20 February 2021).
- Guo, Y.; Liu, Y.; Oerlemans, A.; Lao, S.; Wu, S.; Lew, M.S. Deep learning for visual understanding: A review. Neurocomputing 2016, 187, 27–48. [Google Scholar] [CrossRef]
- Unnikrishnan, A.; Sowmya, V.; Soman, K.P. Deep AlexNet with Reduced Number of Trainable Parameters for Satellite Image Classification. Procedia Comput. Sci. 2018, 143, 931–938. [Google Scholar] [CrossRef]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. Imagenet classification with deep convolutional neural networks. Adv. Neural Inf. Process. Syst. 2012, 25, 1097–1105. [Google Scholar] [CrossRef]
- Donahue, J.; Jia, Y.; Vinyals, O.; Hoffman, J.; Zhang, N.; Tzeng, E.; Darrell, T. DeCAF: A Deep Convolutional Activation Feature for Generic Visual Recognition. In Proceedings of the International Conference on Machine Learning, PMLR, Beijing, China, 21–26 June 2014; pp. 647–655. [Google Scholar]
- Sharif Razavian, A.; Azizpour, H.; Sullivan, J.; Carlsson, S. CNN Features Off-the-Shelf: An Astounding Baseline for Recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition Workshops, Columbus, OH, USA, 24–27 June 2014; pp. 806–813. [Google Scholar]
- Yosinski, J.; Clune, J.; Bengio, Y.; Lipson, H. How transferable are features in deep neural networks? arXiv 2014, arXiv:abs/1411.1792. [Google Scholar]
- Penatti, O.A.B.; Nogueira, K.; dos Santos, J.A. Do Deep Features Generalize From Everyday Objects to Remote Sensing and Aerial Scenes Domains? In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition Workshops, Boston, MA, USA, 7–12 June 2015; pp. 44–51. [Google Scholar]
- Castelluccio, M.; Poggi, G.; Sansone, C.; Verdoliva, L. Land Use Classification in Remote Sensing Images by Convolutional Neural Networks. arXiv 2015, arXiv:abs/1508.00092. [Google Scholar]
- Lyu, H.; Lu, H.; Mou, L. Learning a Transferable Change Rule from a Recurrent Neural Network for Land Cover Change Detection. Remote Sens. 2016, 8, 506. [Google Scholar] [CrossRef] [Green Version]
- Shin, H.C.; Roth, H.R.; Gao, M.; Lu, L.; Xu, Z.; Nogues, I.; Yao, J.; Mollura, D.; Summers, R.M. Deep Convolutional Neural Networks for Computer-Aided Detection: CNN Architectures, Dataset Characteristics and Transfer Learning. IEEE Trans. Med. Imaging 2016, 35, 1285–1298. [Google Scholar] [CrossRef] [Green Version]
- Fu, T.; Ma, L.; Li, M.; Johnson, B.A. Using convolutional neural network to identify irregular segmentation objects from very high-resolution remote sensing imagery. JARS 2018, 12, 025010. [Google Scholar] [CrossRef]
- Nguyen, T.; Han, J.; Park, A.D.C. Satellite image classification using convolutional learning. AIP Conf. Proc. 2013, 1558, 2237–2240. [Google Scholar]
- Marmanis, D.; Datcu, M.; Esch, T.; Stilla, U. Deep Learning Earth Observation Classification Using Image Net Pretrained Networks. IEEE Geosci. Remote Sens. Lett. 2015, 13, 105–109. [Google Scholar] [CrossRef] [Green Version]
- Othman, E.; Bazi, Y.; Alajlan, N.; Alhichri, H.; Melgani, F. Using convolutional features and a sparse autoencoderfor land-use scene classification. Int. J. Remote Sens. 2016, 37, 2149–2167. [Google Scholar] [CrossRef]
- Sharma, A.; Liu, X.; Yang, X.; Shi, D. A patch-based convolutional neural network for remote sensing image classification. Neural Netw. 2017, 95, 19–28. [Google Scholar] [CrossRef] [PubMed]
- Lv, X.; Ming, D.; Lu, T.; Zhou, K.; Wang, M.; Bao, H. A New Method for Region-Based Majority Voting CNNs for Very High Resolution Image Classification. Remote Sens. 2018, 10, 1946. [Google Scholar] [CrossRef] [Green Version]
- CS231n Convolutional Neural Networks for Visual Recognition. Available online: https://cs231n.github.io/convolutional-networks/ (accessed on 31 December 2020).
- Arunava. Convolutional Neural Network—Towards Data Science. Towards Data Science, 25 December 2018. Available online: https://towardsdatascience.com/convolutional-neural-network-17fb77e76c05 (accessed on 31 December 2020).
- Dansbecker. Rectified Linear Units (ReLU) in Deep Learning. Kaggle. 7 May 2018. Available online: https://kaggle.com/dansbecker/rectified-linear-units-relu-in-deep-learning (accessed on 1 April 2021).
- Wood, T. Softmax Function. DeepAI. 17 May 2019. Available online: https://deepai.org/machine-learning-glossary-and-terms/softmax-layer (accessed on 1 April 2021).
- Budhiraja, A. Dropout in (Deep) Machine Learning—Amar Budhiraja—Medium. Medium, 15 December 2016. Available online: https://medium.com/@amarbudhiraja/https-medium-com-amarbudhiraja-learning-less-to-learn-better-dropout-in-deep-machine-learning-74334da4bfc5 (accessed on 31 December 2020).
- Keras Team. Layer Weight Initializers. Available online: https://keras.io/api/layers/initializers/#glorotuniform-class (accessed on 15 February 2021).
- Keras Team. Keras: The Python Deep Learning API. Available online: https://keras.io/ (accessed on 15 February 2021).
- Brownlee, J. A Gentle Introduction to k-Fold Cross-Validation. 22 May 2018. Available online: https://machinelearningmastery.com/k-fold-cross-validation/ (accessed on 31 December 2020).
- Shung, K.P. Accuracy, Precision, Recall or F1?—Towards Data Science. Towards Data Science, 15 March 2018. Available online: https://towardsdatascience.com/accuracy-precision-recall-or-f1-331fb37c5cb9 (accessed on 31 December 2020).















| Types | Code in Annex I to the Habitat Directive and (/) EUNIS Taxonomies | Description |
|---|---|---|
| Type_1 | 6210 (*)/E1.263 where (*) indicates important orchid sites | Semi-natural and natural dry grasslands and scrubland facies on calcareous substrates (Festuco-Brometalia). This habitat in Murgia Alta is limited to small, highly fragmented patches that can be located in areas found at higher quotas, where agriculture and pasture have been abandoned. |
| Type_2 | 62A0/E1.55 | Eastern sub-Mediterranean dry grasslands (Scorzoneratalia villosae). This habitat is the most widespread and dominant habitat in the study area and is characterized by the endemic feather grass Stipa austroitalica, which constitutes perennial prairies with a rocky nature. |
| Type_3 | 6220*/E1.434 where * indicates priority habitat | Pseudo-steppe with grasses and annuals of the Thero-Brachypodietea. In Murgia Alta, this habitat consists of different types of grasslands, both annual and perennial. Annual communities resulting in small patches of less than 10 meters are not considered in the present study. Only Hyparrhenia hirta perennial communities will be considered in this work. |
| Type_4 | No code in Annex I X/E1.61-E1.C2-E1.C4 | Mediterranean subnitrophilous grass communities, thistle fields and giant fennel (Ferula) stands. In the study area, such a grassland type consists of both annual and perennial communities. These grassland communities can be generally found in lower quota areas. Since these areas are easier to access, they have been cultivated and used for sheep grazing. The listed grassland communities include EUNIS taxonomy codes E1.61-E1.C2-E1.C4. |
| Season | Date of Acquisition |
|---|---|
| Winter | 30 January 2018 (biomass pre-peak) |
| Spring | 20 April 2018 (peak of biomass) |
| Summer | 19 July 2018 (dry season) |
| Autumn | 27 October 2018 (biomass post-peak) |
| Layer | Size | Activation Function |
|---|---|---|
| input | patch_size ×patch_size × num_bands (40) | N/A |
| conv | kernel_size × kernel_size, depth (32) | Rectified Linear Unit (ReLU) [66] |
| FC #1 | output_size (128) | ReLU |
| FC #2 | num_classes (4) | softmax [67] |
| # of Patches with Different Sizes | ||||||
|---|---|---|---|---|---|---|
| Class | 1 × 1 | 2 × 2 | 3 × 3 | 4 × 4 | 5 × 5 | 6 × 6 |
| Type_1 | 1.002 | 490 | 193 | 79 | 32 | 13 |
| Type_2 | 65.553 | 58.129 | 51.357 | 45.129 | 39.558 | 34.543 |
| Type_3 | 712 | 543 | 404 | 291 | 207 | 141 |
| Type_4 | 2.221 | 1.365 | 882 | 559 | 348 | 195 |
| Patch Size | OA | Precision | Recall | F1-Score |
|---|---|---|---|---|
| 1 × 1 | 0.965 ± 0.006 | 0.713 | 0.967 | 0.805 |
| 2 × 2 | 0.992 ± 0.001 | 0.884 | 0.967 | 0.923 |
| 3 × 3 | 0.996 ± 0.000 | 0.932 | 0.968 | 0.949 |
| 4 × 4 | 0.997 ± 0.001 | 0.942 | 0.982 | 0.960 |
| 5 × 5 | 0.998 ± 0.001 | 0.960 | 0.982 | 0.971 |
| 6 × 6 | 0.998 ± 0.000 | 0.942 | 0.987 | 0.967 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fazzini, P.; De Felice Proia, G.; Adamo, M.; Blonda, P.; Petracchini, F.; Forte, L.; Tarantino, C. Sentinel-2 Remote Sensed Image Classification with Patchwise Trained ConvNets for Grassland Habitat Discrimination. Remote Sens. 2021, 13, 2276. https://doi.org/10.3390/rs13122276
Fazzini P, De Felice Proia G, Adamo M, Blonda P, Petracchini F, Forte L, Tarantino C. Sentinel-2 Remote Sensed Image Classification with Patchwise Trained ConvNets for Grassland Habitat Discrimination. Remote Sensing. 2021; 13(12):2276. https://doi.org/10.3390/rs13122276
Chicago/Turabian StyleFazzini, Paolo, Giuseppina De Felice Proia, Maria Adamo, Palma Blonda, Francesco Petracchini, Luigi Forte, and Cristina Tarantino. 2021. "Sentinel-2 Remote Sensed Image Classification with Patchwise Trained ConvNets for Grassland Habitat Discrimination" Remote Sensing 13, no. 12: 2276. https://doi.org/10.3390/rs13122276
APA StyleFazzini, P., De Felice Proia, G., Adamo, M., Blonda, P., Petracchini, F., Forte, L., & Tarantino, C. (2021). Sentinel-2 Remote Sensed Image Classification with Patchwise Trained ConvNets for Grassland Habitat Discrimination. Remote Sensing, 13(12), 2276. https://doi.org/10.3390/rs13122276