Forest Leaf Mass per Area (LMA) through the Eye of Optical Remote Sensing: A Review and Future Outlook
Abstract
:1. Introduction
2. LMA Chemical Composition and Variation over Space and Time
3. Remote Sensing of Forest LMA and Its Scaling

| Wavelength (nm) | Electron Transition | Absorbing Compound |
|---|---|---|
| 910 | C-H stretch, 3rd overtone | Protein |
| 970 | O-H bend, 1st overtone | Starch |
| 990 | O-H stretch, 2nd overtone | Starch |
| 1020 | N-H stretch | Protein |
| 1120 | C-H stretch, 2nd overtone | Lignin |
| 1200 | O-H bend, 1st overtone | Cellulose, starch, lignin |
| 1420 | C-H stretch, C-H deformation | Lignin |
| 1450 | O-H stretch, 1st overtone C-H stretch, C-H deformation | Starch, sugar, lignin |
| 1490 | O-H stretch, 1st overtone | Cellulose, sugar |
| 1510 | N-H stretch, 1st overtone | Protein, nitrogen |
| 1530 | O-H stretch, 1st overtone | Starch |
| 1540 | O-H stretch, 1st overtone | Starch, cellulose |
| 1580 | O-H stretch, 1st overtone | Starch, sugar |
| 1690 | C-H stretch, 1st overtone | Lignin, starch, protein, nitrogen |
| 1730 | C-H-stretch | Protein |
| 1736 | O-H stretch | Cellulose |
| 1780 | C-H stretch, 1st overtone O-H stretch H-O-H deformation | Cellulose, sugar, starch |
| 1820 | O-H stretch, C-O stretch, 2nd overtone | Cellulose |
| 1900 | O-H stretch, C-O stretch | Starch |
| 1924 | O-H stretch, O-H deformation | Cellulose |
| 1940 | O-H stretch, O-H deformation | Water, lignin, protein, nitrogen, starch, cellulose |
| 1960 | O-H stretch, O-H bend | Sugar, starch |
| 1980 | N-H asymmetry | Protein |
| 2000 | O-H deformation C-O deformation | Starch |
| 2060 | N=H bend, 2nd overtone N=H bend N-H stretch | Protein, nitrogen |
| 2080 | O-H stretch, O- deformation | Sugar, starch |
| 2100 | O-H bend/ C-O stretch C-O-C stretch, 3rd overtone | Starch cellulose |
| 2130 | N-H stretch | Protein |
| 2180 | N-H bend, 2nd overtone, C-H stretch, C-O stretch, C=O stretch C-N stretch | Protein, nitrogen |
| 2240 | C-H stretch | Protein |
| 2250 | O-H stretch, O-H deformation | Starch |
| 2270 | C-H stretch, O-H stretch, CH2 bend, CH2 stretch | Cellulose, sugar, starch |
| 2280 | C-H stretch, CH2 deformation | Starch, Cellulose |
| 2300 | N-H stretch, C=O stretch, C-H bend, 2nd overtone | Protein, nitrogen |
| 2310 | C-H bend, 2nd overtone | Oil |
| 2320 | C-H stretch, CH2 deformation | Starch |
| 2340 | C-H deformation, O-H deformation, C-H deformation, O-H deformation | Cellulose |
| 2350 | CH2 bend, 2nd overtone, C-H deformation, 2nd overtone | Cellulose, protein, nitrogen |
4. Remote Sensing Systems in LMA Retrieval and Estimation
4.1. Near-Ground Based Platforms
4.2. Airborne Platforms
4.3. Spaceborne Satellite Platforms
4.4. Challenges in the Estimation of LMA using Air- and Space-borne Systems
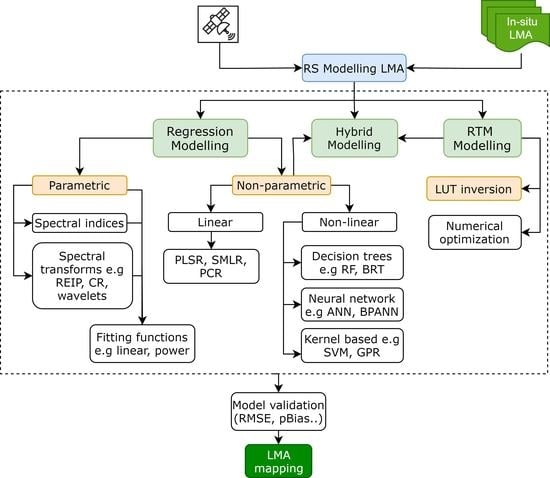
5. Models to Estimate/Retrieve LMA from Remote Sensing Data
5.1. Statistical Modelling
5.2. Physical Models
5.3. Hybrid Modelling
5.4. Challenges in Remote Sensing Modeling of LMA
| Category | Method | Spectral Data | Sensor | Scale | Main Findings Reported | Reference |
|---|---|---|---|---|---|---|
| Parametric regressions | Continuous wavelet transform | Leaf hyperspectral reflectance | Field spectrometer | leaf | Wavelet features at 1639 nm and 2133 nm, yielded the most accurate model to estimate LMA (R2 = 0.74, RMSE = 18.97 g m−2) | [63] |
| Non-parametric linear regressions | PLS | In situ leaf reflectance | Field spectrometer | leaf | A multibiome leaf spectra–LMA PLS model was built explaining 85% variance in LMA. The model incorporating vegetation from the Arctic to the tropics, included broad- and needle leaf species, sunlit and shade foliar yielded a RMSE of 15.45 g m−2 | [26] |
| PLS | In situ optical and thermal reflectance | Field spectrometer | leaf | Synergy of Visible Short Wave Infrared (VSWIR) and Thermal Infrared spectrum (TIR) improve LMA prediction (RMSEP = 18.31) compared to using the spectral regions in isolation | [117] | |
| SMLR | Leaf reflectance and derive spectra | Field spectrometer | leaf | Wavebands selected by the SMLR did not match known absorption features of LMA and other related traits. The SMLR performed differently depending on the expression used i.e., more accurate models were generated using content (g m−2) compared to concentration (g g−1) | [114] | |
| PLS | Airborne hyperspectral | Carnegie Airborne Observatory | canopy | VSWIR and LiDAR generated R2 = 0.69 and RMSE = 9.99% in LMA estimation | [32] | |
| PLS | Airborne hyperspectral and LiDAR | NEON’s Airborne Observatory Platform- AVIRIS-NG-like sensor | canopy | Combining top-of-canopy (R2 = 0.57, RMSE = 10.8 g m−2) and within canopy (R2 = 0.78, RMSE = 8.3 g m−2) LMA, significantly improved three-dimensional PLSR modelling (R2 = 0.82, RMSE = 8.5 g m−2) of LMA. The 2000–2450 nm spectral subset generated the highest accuracy (%RMSE = 15.37) compared to the other spectral subsets (400–2450, 800–2450, 1600–2450 nm) | [62] | |
| Non-linear parametric regressions | SVM | In situ leaf reflectance | Field spectrometer | leaf | SVM using spectral data between 900-2400 nm generated a RMSE of 2.52 mg cm−2 | [59] |
| RF | Raw bands and spectral indices | Sentinel-2 | canopy | LMA varied significantly (p < 0.05) across the canopy between sunlit and shaded. A weighted canopy expression outperformed (R2 = 0.67, NRMSE = 0.16) the traditional sunlit based expression (R2 = 0.54, NRMSE = 0.18). predictive maps of LMA were generated using Sentinel-2 bands and vegetation indices. | [44] | |
| Physical models (RTM based) | PROSPECT-PRO | Leaf reflectance and transmittance | Field spectrometer | leaf | PROSPECT PRO separates LMA into the nitrogen-based constituents (proteins) and CBC (carbon-based constituents i.e., cellulose, lignin, hemicellulose, starch, and sugars) CBC was accurately estimated for both fresh (R2 = 0.96, NRMSE = 9.6%) and dry leaves samples (R2 = 0.95 and 13.4%) while the sum of CBC and proteins (LMA) was estimated (R2 = 0.90 and NRMSE = 0.165) | [22] |
| PROSPECT | Leaf reflectance | Field spectrometer | leaf | LMA across the vertical canopy profile throughout the growing season was successfully retrieved (R2 0.54–0.82, NRMSE 0.15–0.24) from PROSPECT simulations using the LUT inversion. The best retrieval was obtained for the summer (R2 = 0.82, NRMSE= 0.15) and for upper canopy leaf samples (R2 = 0.61 NRMSE = 0.15) | [24] | |
| PROSAIL | Airborne hyperspectral | AVIRIS | canopy | PROSAIL inversion yielded a RMSE of 0.004. | [21] | |
| Hybrid models | PROSPECT and PLS | Leaf reflectance | Field spectrometer | leaf | A PLS model calibrated using PROSPECT-5 spectral simulations yielded an RMSE of 0.007 g cm−2 on experimental data compared to spectral index (NDLMA= 0.0021 g cm−2) | [23] |
| Spectral indices | Leaf hyperspectral reflectance | Field spectrometer | leaf | A narrow band index (normalized dry matter index, NDMI) centered at 1649 and 1722 nm developed from PROSPECT simulations (R2 = 0.85 RMSE 0.0019 g cm−2 and validated on the LOPEX dataset (R2 = 0.68, RMSE = 0.0014 g cm−2) yielded the lowest estimation error | [18] |
6. Research Gaps and Future Outlook
7. Conclusions
- Studies on remote sensing of LMA are mainly based on leaf reflectance measured using field spectrometers. A number of studies have been conducted using airborne and spaceborne sensors. With the availability of multispectral sensors, such as Sentinel-2 and Landsat-8, and new generation sensors, such as WorldView and GeoEye, further research is required to assess the utility of these sensors to characterize a key EBV at a large spatial scale.
- Most studies on the estimation/retrieval of LMA have been conducted using the optical range of 400–2500 nm. A few studies have assessed the utility of sensor integration, especially data in the thermal spectrum for LMA estimation. Upcoming sensors such as HyspIRI, which sense radiance in the thermal domain, will provide an opportunity to test and upscale LMA estimation in the thermal domain over large spatial extents.
- Optical imagery can be used to estimate LMA in two-dimensional space. Studies have demonstrated that LMA significantly varies across the canopy vertical profile due to variation in radiance. Therefore, the characterization of LMA in three-dimensional space by synergizing optical sensors and LiDAR products requires further investigation in different forest types at various temporal domains.
- Despite recent achievements in the separation of LMA constituents in radiative models such as PROSPECT, continuous efforts to unbundle LMA constituents remain an ongoing process. The modified PROSPECT models require further testing by scaling them to canopy and landscape scale in forest ecosystems.
- The advancement in novel non-parametric algorithms, such as GPR, and the improvement in physical models, such as PROSPECT PRO, have provided opportunities for validating the utility of hybrid models in LMA retrieval from RTM simulations. Currently, hybrid models for LMA retrieval have been calibrated based on earlier versions of radiative transfer models and non-parametric models such as PLS.
- There is potential confusion regarding the terminology used in scientific reports in referring to LMA. Terms such as mass-based leaf dry matter content (LDMC) and specific leaf weight have been used interchangeably with LMA. Consistent use of the term LMA to refer to the area-based dry matter is encouraged.
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- He, L.; Chen, J.; Pan, Y.; Birdsey, R.; Kattge, J. Relationships between net primary productivity and forest stand age in U.S. Forests. Glob. Biogeochem. Cycles 2012, 26, 1–16. [Google Scholar] [CrossRef]
- Green, D.; Erickson, J.; Kruger, E. Foliar morphology and canopy nitrogen as predictors of light-use efficiency in terrestrial vegetation. Agric. For. Meteorol. 2003, 115, 163–171. [Google Scholar] [CrossRef]
- Poorter, H.; Niinemets, Ü.; Poorter, L.; Wright, I.J.; Villar, R. Causes and consequences of variation in leaf mass per area (LMA): A meta-analysis. New Phytol. 2009, 182, 565–588. [Google Scholar] [CrossRef] [PubMed]
- Pereira, H.M.; Ferrier, S.; Walters, M.; Geller, G.N.; Jongman, R.H.G.; Scholes, R.J.; Bruford, M.W.; Brummitt, N.; Butchart, S.H.M.; Cardoso, A.C.; et al. Essential biodiversity variables. Science 2013, 339, 277. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jetz, W.; McGeoch, M.A.; Guralnick, R.; Ferrier, S.; Beck, J.; Costello, M.J.; Fernandez, M.; Geller, G.N.; Keil, P.; Merow, C.; et al. Essential biodiversity variables for mapping and monitoring species populations. Nat. Ecol. Evol. 2019, 3, 539–551. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qin, J.; Shangguan, Z. Effects of forest types on leaf functional traits and their interrelationships of pinus massoniana coniferous and broad-leaved mixed forests in the subtropical mountain, southeastern China. Ecol. Evol. 2019, 9, 6922–6932. [Google Scholar] [CrossRef] [Green Version]
- Wright, I.J.; Reich, P.B.; Westoby, M.; Ackerly, D.D.; Baruch, Z.; Bongers, F.; Cavender-Bares, J.; Chapin, T.; Cornelissen, J.H.C.; Diemer, M.; et al. The worldwide leaf economics spectrum. Nature 2004, 428, 821–827. [Google Scholar] [CrossRef]
- Oren, R.; Schulze, E.D.; Matyssek, R.; Zimmermann, R. Estimating photosynthetic rate and annual carbon gain in conifers from specific leaf weight and leaf biomass. Oecologia 1986, 70, 187–193. [Google Scholar] [CrossRef]
- Villar, R.; Ruiz-Robleto, J.; Ubera, J.L.; Poorter, H. Exploring variation in leaf mass per area (LMA) from leaf to cell: An anatomical analysis of 26 woody species. Am. J. Bot. 2013, 100, 1969–1980. [Google Scholar] [CrossRef]
- Abdullah, H.; Darvishzadeh, R.; Skidmore, A.K.; Groen, T.A.; Heurich, M. European spruce bark beetle (ips typographus, l.) green attack affects foliar reflectance and biochemical properties. Int. J. Appl. Earth Obs. Geoinf. 2018, 64, 199–209. [Google Scholar] [CrossRef] [Green Version]
- Kattge, J.; Díaz, S.; Lavorel, S.; Prentice, I.C.; Leadley, P.; Bönisch, G.; Garnier, E.; Westoby, M.; Reich, P.B.; Wright, I.J.; et al. Try—A global database of plant traits. Glob. Change Biol. 2011, 17, 2905–2935. [Google Scholar] [CrossRef]
- Homolová, L.; Malenovský, Z.; Clevers, J.G.P.W.; García-Santos, G.; Schaepman, M.E. Review of optical-based remote sensing for plant trait mapping. Ecol. Complex. 2013, 15, 1–16. [Google Scholar] [CrossRef] [Green Version]
- Rahimzadeh-Bajgiran, P.; Hennigar, C.; Weiskittel, A.; Lamb, S. Forest potential productivity mapping by linking remote-sensing-derived metrics to site variables. Remote Sens. 2020, 12, 2056. [Google Scholar] [CrossRef]
- Shull, C.A. A spectrophotometric study of reflection of light from leaf surfaces. Bot. Gaz. 1929, 87, 583–607. [Google Scholar] [CrossRef]
- Le Maire, G.; François, C.; Soudani, K.; Berveiller, D.; Pontailler, J.-Y.; Bréda, N.; Genet, H.; Davi, H.; Dufrêne, E. Calibration and validation of hyperspectral indices for the estimation of broadleaved forest leaf chlorophyll content, leaf mass per area, leaf area index and leaf canopy biomass. Remote Sens. Environ. 2008, 112, 3846–3864. [Google Scholar] [CrossRef]
- Romero, A.; Aguado, I.; Yebra, M. Estimation of dry matter content in leaves using normalized indexes and prospect model inversion. Int. J. Remote Sens. 2012, 33, 396–414. [Google Scholar] [CrossRef]
- Yang, X.; Tang, J.; Mustard, J.F.; Wu, J.; Zhao, K.; Serbin, S.; Lee, J.-E. Seasonal variability of multiple leaf traits captured by leaf spectroscopy at two temperate deciduous forests. Remote Sens. Environ. 2016, 179, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Qu, J.J.; Hao, X.; Hunt, E.R. Estimating dry matter content from spectral reflectance for green leaves of different species. Int. J. Remote Sens. 2011, 32, 7097–7109. [Google Scholar] [CrossRef]
- Jacquemoud, S.; Baret, F. Prospect: A model of leaf optical properties spectra. Remote Sens. Environ. 1990, 34, 75–91. [Google Scholar] [CrossRef]
- Jacquemoud, S.; Verhoef, W.; Baret, F.; Bacour, C.; Zarco-Tejada, P.J.; Asner, G.P.; François, C.; Ustin, S.L. PROSPECT + SAIL models: A review of use for vegetation characterization. Remote Sens. Environ. 2009, 113 (Suppl. 1), S56–S66. [Google Scholar] [CrossRef]
- Casas, A.; Riaño, D.; Ustin, S.L.; Dennison, P.; Salas, J. Estimation of water-related biochemical and biophysical vegetation properties using multitemporal airborne hyperspectral data and its comparison to MODIS spectral response. Remote Sens. Environ. 2014, 148, 28–41. [Google Scholar] [CrossRef]
- Féret, J.-B.; Berger, K.; de Boissieu, F.; Malenovský, Z. PROSPECT-PRO for estimating content of nitrogen-containing leaf proteins and other carbon-based constituents. Remote Sens. Environ. 2021, 252, 112173. [Google Scholar] [CrossRef]
- Féret, J.-B.; François, C.; Gitelson, A.; Asner, G.P.; Barry, K.M.; Panigada, C.; Richardson, A.D.; Jacquemoud, S. Optimizing spectral indices and chemometric analysis of leaf chemical properties using radiative transfer modeling. Remote Sens. Environ. 2011, 115, 2742–2750. [Google Scholar] [CrossRef] [Green Version]
- Gara, T.W.; Darvishzadeh, R.; Skidmore, A.K.; Wang, T.; Heurich, M. Evaluating the performance of PROSPECT in the retrieval of leaf traits across canopy throughout the growing season. Int. J. Appl. Earth Obs. Geoinf. 2019, 83, 101919. [Google Scholar] [CrossRef]
- Berger, K.; Atzberger, C.; Danner, M.; D’Urso, G.; Mauser, W.; Vuolo, F.; Hank, T. Evaluation of the PROSAIL model capabilities for future hyperspectral model environments: A review study. Remote Sens. 2018, 10, 85. [Google Scholar] [CrossRef] [Green Version]
- Serbin, S.P.; Wu, J.; Ely, K.S.; Kruger, E.L.; Townsend, P.A.; Meng, R.; Wolfe, B.T.; Chlus, A.; Wang, Z.; Rogers, A. From the arctic to the tropics: Multibiome prediction of leaf mass per area using leaf reflectance. New Phytol. 2019, 224, 1557–1568. [Google Scholar] [CrossRef]
- Serbin, S.P.; Townsend, P.A. Scaling functional traits from leaves to canopies. In Remote Sensing of Plant Biodiversity; Cavender-Bares, J., Gamon, J.A., Townsend, P.A., Eds.; Springer: Cham, Switzerland, 2020; pp. 43–82. [Google Scholar]
- Verrelst, J.; Camps-Valls, G.; Muñoz-Marí, J.; Rivera, J.P.; Veroustraete, F.; Clevers, J.G.P.W.; Moreno, J. Optical remote sensing and the retrieval of terrestrial vegetation bio-geophysical properties—A review. ISPRS J. Photogramm. Remote Sens. 2015, 108, 273–290. [Google Scholar] [CrossRef]
- Verrelst, J.; Malenovský, Z.; Van der Tol, C.; Camps-Valls, G.; Gastellu-Etchegorry, J.-P.; Lewis, P.; North, P.; Moreno, J. Quantifying vegetation biophysical variables from imaging spectroscopy data: A review on retrieval methods. Surv. Geophys. 2019, 40, 589–629. [Google Scholar] [CrossRef] [Green Version]
- Houborg, R.; Fisher, J.B.; Skidmore, A.K. Advances in remote sensing of vegetation function and traits. Int. J. Appl. Earth Obs. Geoinf. 2015, 43, 1–6. [Google Scholar] [CrossRef] [Green Version]
- Gutschick, V.P.; Wiegel, F.W. Optimizing the canopy photosynthetic rate by patterns of investment in specific leaf mass. Am. Nat. 1988, 132, 67–86. [Google Scholar] [CrossRef]
- Asner, G.P.; Knapp, D.E.; Anderson, C.B.; Martin, R.E.; Vaughn, N. Large-scale climatic and geophysical controls on the leaf economics spectrum. Proc. Natl. Acad. Sci. USA 2016, 113, E4043. [Google Scholar] [CrossRef] [Green Version]
- McGill, B.J.; Enquist, B.J.; Weiher, E.; Westoby, M. Rebuilding community ecology from functional traits. Trends Ecol. Evol. 2006, 21, 178–185. [Google Scholar] [CrossRef]
- Raymond Hunt, J.E.; Lingli, W.; John, J.Q.; Xianjun, H. Remote sensing of fuel moisture content from canopy water indices and normalized dry matter index. APPRES 2012, 6, 1–11. [Google Scholar] [CrossRef]
- Wang, L.; Hunt, E.R.; Qu, J.J.; Hao, X.; Daughtry, C.S.T. Remote sensing of fuel moisture content from ratios of narrow-band vegetation water and dry-matter indices. Remote Sens. Environ. 2013, 129, 103–110. [Google Scholar] [CrossRef] [Green Version]
- Asner, G.P.; Martin, R.; Tupayachi, R.; Emerson, R.; Martinez, P.; Sinca, F.N.; Powell, G.V.N.; Wright, S.J.; Lugo, A.E. Taxonomy and remote sensing of leaf mass per area (LMA) in humid tropical forests. Ecol. Appl. 2011, 21, 85–98. [Google Scholar] [CrossRef]
- Witkowski, E.T.F.; Lamont, B.B. Leaf specific mass confounds leaf density and thickness. Oecologia 1991, 88, 486–493. [Google Scholar] [CrossRef]
- Niinemets, Ü. Research review. Components of leaf dry mass per area—Thickness and density—Alter leaf photosynthetic capacity in reverse directions in woody plants. New Phytol. 1999, 144, 35–47. [Google Scholar] [CrossRef]
- Baret, F.; Fourty, T. Estimation of leaf water content and specific leaf weight from reflectance and transmittance measurements. Agronomie 1997, 17, 455–464. [Google Scholar] [CrossRef] [Green Version]
- Sims, D.A.; Gamon, J.A. Relationships between leaf pigment content and spectral reflectance across a wide range of species, leaf structures and developmental stages. Remote Sens. Environ. 2002, 81, 337–354. [Google Scholar] [CrossRef]
- Fourty, T.; Baret, F. On spectral estimates of fresh leaf biochemistry. Int. J. Remote Sens. 1998, 19, 1283–1297. [Google Scholar] [CrossRef]
- Lee, M.A. A global comparison of the nutritive values of forage plants grown in contrasting environments. J. Plant Res. 2018, 131, 641–654. [Google Scholar] [CrossRef]
- Wang, Z.; Chlus, A.; Geygan, R.; Ye, Z.; Zheng, T.; Singh, A.; Couture, J.J.; Cavender-Bares, J.; Kruger, E.L.; Townsend, P.A. Foliar functional traits from imaging spectroscopy across biomes in eastern north America. New Phytol. 2020, 228, 494–511. [Google Scholar] [CrossRef] [PubMed]
- Gara, T.W.; Darvishzadeh, R.; Skidmore, A.K.; Wang, T.; Heurich, M. Accurate modelling of canopy traits from seasonal Sentinel-2 imagery based on the vertical distribution of leaf traits. ISPRS J. Photogramm. Remote Sens. 2019, 157, 108–123. [Google Scholar] [CrossRef]
- Shipley, B. Structured interspecific determinants of specific leaf area in 34 species of herbaceous angiosperms. Funct. Ecol. 1995, 9, 312–319. [Google Scholar] [CrossRef]
- Chen, J.-L.; Reynolds, J.F.; Harley, P.C.; Tenhunen, J.D. Coordination theory of leaf nitrogen distribution in a canopy. Oecologia 1993, 93, 63–69. [Google Scholar] [CrossRef] [PubMed]
- Hirose, T.; Werger, M.J.A. Maximizing daily canopy photosynthesis with respect to the leaf nitrogen allocation pattern in the canopy. Oecologia 1987, 72, 520–526. [Google Scholar] [CrossRef] [PubMed]
- Aranda, I.; Pardo, F.; Gil, L.; Pardos, J.A. Anatomical basis of the change in leaf mass per area and nitrogen investment with relative irradiance within the canopy of eight temperate tree species. Acta Oecologica 2004, 25, 187–195. [Google Scholar] [CrossRef]
- Li, H.; Zhao, C.; Huang, W.; Yang, G. Non-uniform vertical nitrogen distribution within plant canopy and its estimation by remote sensing: A review. Field Crop. Res. 2013, 142, 75–84. [Google Scholar] [CrossRef]
- Wright, I.J.; Leishman, M.R.; Read, C.; Westoby, M. Gradients of light availability and leaf traits with leaf age and canopy position in 28 Australian shrubs and trees. Funct. Plant Biol. 2006, 33, 407–419. [Google Scholar] [CrossRef] [Green Version]
- Moreno-Martínez, Á.; Camps-Valls, G.; Kattge, J.; Robinson, N.; Reichstein, M.; van Bodegom, P.; Kramer, K.; Cornelissen, J.H.C.; Reich, P.; Bahn, M.; et al. A methodology to derive global maps of leaf traits using remote sensing and climate data. Remote Sens. Environ. 2018, 218, 69–88. [Google Scholar] [CrossRef] [Green Version]
- Chaves, M.; Marôco, J.; Pereira, J. Understanding plant responses to drought—from genes to the whole plant. Funct. Plant Biol. FPB 2003, 30, 239–264. [Google Scholar] [CrossRef] [PubMed]
- Lambers, H.; Poorter, H. Inherent variation in growth rate between higher plants: A search for physiological causes and ecological consequences. In Advances in Ecological Research; Begon, M., Fitter, A.H., Eds.; Academic Press: Cambridge, MA, USA, 1992; Volume 23, pp. 187–261. [Google Scholar]
- Li, L.; Wang, X.; Niu, J.; Cui, J.; Zhang, Q.; Wan, W.; Liu, B. Effects of elevated atmospheric O3 concentrations on early and late leaf growth and elemental contents of acer truncatum bung under mild drought. Acta Ecol. Sin. 2017, 37, 31–34. [Google Scholar] [CrossRef]
- Pakeman, R.J.; Quested, H.M. Sampling plant functional traits: What proportion of the species need to be measured? Appl. Veg. Sci. 2007, 10, 91–96. [Google Scholar] [CrossRef]
- Kumar, L.; Schmidt, K.; Dury, S.; Skidmore, A. Imaging spectrometry and vegetation science. In Imaging Spectrometry: Basic Principles and Prospective Applications; Meer, F.D., Jong, S.M.D., Eds.; Springer: Dordrecht, The Netherlands, 2001; pp. 111–155. [Google Scholar]
- Curran, P.J. Remote sensing of foliar chemistry. Remote Sens. Environ. 1989, 30, 271–278. [Google Scholar] [CrossRef]
- Qiu, F.; Chen, J.M.; Ju, W.; Wang, J.; Zhang, Q.; Fang, M. Improving the prospect model to consider anisotropic scattering of leaf internal materials and its use for retrieving leaf biomass in fresh leaves. IEEE Trans. Geosci. Remote Sens. 2018, 56, 3119–3136. [Google Scholar] [CrossRef]
- Féret, J.B.; le Maire, G.; Jay, S.; Berveiller, D.; Bendoula, R.; Hmimina, G.; Cheraiet, A.; Oliveira, J.C.; Ponzoni, F.J.; Solanki, T.; et al. Estimating leaf mass per area and equivalent water thickness based on leaf optical properties: Potential and limitations of physical modeling and machine learning. Remote Sens. Environ. 2019, 231, 110959. [Google Scholar] [CrossRef]
- Riano, D.; Vaughan, P.; Chuvieco, E.; Zarco-Tejada, P.J.; Ustin, S.L. Estimation of fuel moisture content by inversion of radiative transfer models to simulate equivalent water thickness and dry matter content: Analysis at leaf and canopy level. IEEE Trans. Geosci. Remote. Sens. 2005, 43, 819–826. [Google Scholar] [CrossRef]
- Zhao, K.; Valle, D.; Popescu, S.; Zhang, X.; Mallick, B. Hyperspectral remote sensing of plant biochemistry using bayesian model averaging with variable and band selection. Remote Sens. Environ. 2013, 132, 102–119. [Google Scholar] [CrossRef]
- Chlus, A.; Kruger, E.L.; Townsend, P.A. Mapping three-dimensional variation in leaf mass per area with imaging spectroscopy and lidar in a temperate broadleaf forest. Remote Sens. Environ. 2020, 250, 112043. [Google Scholar] [CrossRef]
- Cheng, T.; Rivard, B.; Sánchez-Azofeifa, A.G.; Féret, J.-B.; Jacquemoud, S.; Ustin, S.L. Deriving leaf mass per area (LMA) from foliar reflectance across a variety of plant species using continuous wavelet analysis. ISPRS J. Photogramm. Remote Sens. 2014, 87, 28–38. [Google Scholar] [CrossRef]
- Ali, A.M.; Darvishzadeh, R.; Skidmore, A.K.; Duren, I.v. Effects of canopy structural variables on retrieval of leaf dry matter content and specific leaf area from remotely sensed data. Sel. Top. Appl. Earth Obs. Remote Sens. IEEE J. 2015, 9, 898–909. [Google Scholar] [CrossRef]
- Ollinger, S.V. Sources of variability in canopy reflectance and the convergent properties of plants. New Phytol. 2011, 189, 375–394. [Google Scholar] [CrossRef]
- Townsend, P.A.; Foster, J.R.; Chastain, R.A.; Currie, W.S. Application of imaging spectroscopy to mapping canopy nitrogen in the forests of the central appalachian mountains using Hyperion and AVIRIS. IEEE Trans. Geosci. Remote Sens. 2003, 41, 1347–1354. [Google Scholar] [CrossRef]
- Asner, G.P.; Martin, R.E.; Anderson, C.B.; Knapp, D.E. Quantifying forest canopy traits: Imaging spectroscopy versus field survey. Remote Sens. Environ. 2015, 158, 15–27. [Google Scholar] [CrossRef]
- Gara, T.W. Quantitative Remote Sensing of Essential Biodiversity Variables. Ph.D. Thesis, University of Twente, ITC, Enschede, The Netherlands, 2019. [Google Scholar]
- Gara, T.W.; Skidmore, A.K.; Darvishzadeh, R.; Wang, T. Leaf to canopy upscaling approach affects the estimation of canopy traits. GISci. Remote Sens. 2019, 56, 554–575. [Google Scholar]
- Lavorel, S.; Grigulis, K.; McIntyre, S.; Williams, N.S.G.; Garden, D.; Dorrough, J.; Berman, S.; Quétier, F.; Thébault, A.; Bonis, A. Assessing functional diversity in the field—Methodology matters! Funct. Ecol. 2008, 22, 134–147. [Google Scholar] [CrossRef]
- Asner, G.P.; Martin, R.E.; Anderson, C.B.; Kryston, K.; Vaughn, N.; Knapp, D.E.; Bentley, L.P.; Shenkin, A.; Salinas, N.; Sinca, F.; et al. Scale dependence of canopy trait distributions along a tropical forest elevation gradient. New Phytol. 2016, 214, 973–988. [Google Scholar] [CrossRef] [PubMed]
- Nunes, M.H.; Davey, M.P.; Coomes, D.A. On the challenges of using field spectroscopy to measure the impact of soil type on leaf traits. Biogeosciences 2017, 14, 3371–3385. [Google Scholar] [CrossRef] [Green Version]
- Milton, E.J.; Schaepman, M.E.; Anderson, K.; Kneubühler, M.; Fox, N. Progress in field spectroscopy. Remote Sens. Environ. 2009, 113, S92–S109. [Google Scholar] [CrossRef] [Green Version]
- Thome, K.; Wenny, B.; Anderson, N.; McCorkel, J.; Czapla-Myers, J.; Biggar, S. Ultra-portable field transfer radiometer for vicarious calibration of earth imaging sensors. Metrologia 2018, 55, S104–S117. [Google Scholar] [CrossRef] [Green Version]
- Le Maire, G.; François, C.; Dufrêne, E. Towards universal broad leaf chlorophyll indices using prospect simulated database and hyperspectral reflectance measurements. Remote Sens. Environ. 2004, 89, 1–28. [Google Scholar] [CrossRef]
- Li, P.; Wang, Q. Developing and validating novel hyperspectral indices for leaf area index estimation: Effect of canopy vertical heterogeneity. Ecol. Indic. 2013, 32, 123–130. [Google Scholar] [CrossRef]
- Wang, Q.; Li, P. Hyperspectral indices for estimating leaf biochemical properties in temperate deciduous forests: Comparison of simulated and measured reflectance data sets. Ecol. Indic. 2012, 14, 56–65. [Google Scholar] [CrossRef]
- Cho, M.A.; Skidmore, A.K. A new technique for extracting the red edge position from hyperspectral data: The linear extrapolation method. Remote Sens. Environ. 2006, 101, 181–193. [Google Scholar] [CrossRef]
- Wang, Z.; Skidmore, A.K.; Wang, T.; Darvishzadeh, R.; Hearne, J. Applicability of the prospect model for estimating protein and cellulose + lignin in fresh leaves. Remote Sens. Environ. 2015, 168, 205–218. [Google Scholar] [CrossRef]
- Schlerf, M.; Atzberger, C. Inversion of a forest reflectance model to estimate structural canopy variables from hyperspectral remote sensing data. Remote Sens. Environ. 2006, 100, 281–294. [Google Scholar] [CrossRef]
- Wang, Z.; Skidmore, A.K.; Darvishzadeh, R.; Wang, T. Mapping forest canopy nitrogen content by inversion of coupled leaf-canopy radiative transfer models from airborne hyperspectral imagery. Agric. For. Meteorol. 2018, 253–254, 247–260. [Google Scholar] [CrossRef]
- Féret, J.B.; Gitelson, A.A.; Noble, S.D.; Jacquemoud, S. PROSPECT-D: Towards modeling leaf optical properties through a complete lifecycle. Remote Sens. Environ. 2017, 193, 204–215. [Google Scholar] [CrossRef] [Green Version]
- Hoeppner, J.M.; Skidmore, A.K.; Darvishzadeh, R.; Heurich, M.; Chang, H.-C.; Gara, T.W. Mapping canopy chlorophyll content in a temperate forest using airborne hyperspectral data. Remote Sens. 2020, 12, 3573. [Google Scholar] [CrossRef]
- Blackburn, G.A. Remote sensing of forest pigments using airborne imaging spectrometer and lidar imagery. Remote Sens. Environ. 2002, 82, 311–321. [Google Scholar] [CrossRef]
- Singh, A.; Serbin, S.P.; McNeil, B.E.; Kingdon, C.C.; Townsend, P.A. Imaging spectroscopy algorithms for mapping canopy foliar chemical and morphological traits and their uncertainties. Ecol. Appl. 2015, 25, 2180–2197. [Google Scholar] [CrossRef] [Green Version]
- Chadwick, K.D.; Asner, G.P. Organismic-scale remote sensing of canopy foliar traits in lowland tropical forests. Remote Sens. 2016, 8, 87. [Google Scholar] [CrossRef] [Green Version]
- Chauhan, S.; Darvishzadeh, R.; Boschetti, M.; Pepe, M.; Nelson, A. Remote sensing-based crop lodging assessment: Current status and perspectives. ISPRS J. Photogramm. Remote Sens. 2019, 151, 124–140. [Google Scholar] [CrossRef] [Green Version]
- Xie, C.; Yang, C. A review on plant high-throughput phenotyping traits using uav-based sensors. Comput. Electron. Agric. 2020, 178, 105731. [Google Scholar] [CrossRef]
- Thomson, E.R.; Malhi, Y.; Bartholomeus, H.; Oliveras, I.; Gvozdevaite, A.; Peprah, T.; Suomalainen, J.; Quansah, J.; Seidu, J.; Adonteng, C.; et al. Mapping the leaf economic spectrum across west african tropical forests using UAV-acquired hyperspectral imagery. Remote Sens. 2018, 10, 1532. [Google Scholar] [CrossRef] [Green Version]
- Wallis, C.I.B.; Homeier, J.; Peña, J.; Brandl, R.; Farwig, N.; Bendix, J. Modeling tropical montane forest biomass, productivity and canopy traits with multispectral remote sensing data. Remote Sens. Environ. 2019, 225, 77–92. [Google Scholar] [CrossRef]
- Shamsoddini, A.; Raval, S. Mapping red edge-based vegetation health indicators using landsat tm data for Australian native vegetation cover. Earth Sci. Inform. 2018, 11, 545–552. [Google Scholar] [CrossRef]
- Coops, N.C.; Smith, M.L.; Martin, M.E.; Ollinger, S.V. Prediction of eucalypt foliage nitrogen content from satellite-derived hyperspectral data. IEEE Trans. Geosci. Remote. Sens. 2003, 41, 1338–1346. [Google Scholar] [CrossRef]
- Lee, C.M.; Cable, M.L.; Hook, S.J.; Green, R.O.; Ustin, S.L.; Mandl, D.J.; Middleton, E.M. An introduction to the NASA hyperspectral infrared imager (HYSPIRI) mission and preparatory activities. Remote Sens. Environ. 2015, 167, 6–19. [Google Scholar] [CrossRef]
- Shoko, C.; Mutanga, O.; Dube, T. Progress in the remote sensing of C3 and C4 grass species aboveground biomass over time and space. ISPRS J. Photogramm. Remote. Sens. 2016, 120, 13–24. [Google Scholar] [CrossRef]
- Le Maire, G.; Marsden, C.; Verhoef, W.; Ponzoni, F.J.; Lo Seen, D.; Bégué, A.; Stape, J.-L.; Nouvellon, Y. Leaf area index estimation with MODIS reflectance time series and model inversion during full rotations of eucalyptus plantations. Remote Sens. Environ. 2011, 115, 586–599. [Google Scholar] [CrossRef]
- Verrelst, J.; Rivera, J.P.; Veroustraete, F.; Muñoz-Marí, J.; Clevers, J.G.P.W.; Camps-Valls, G.; Moreno, J. Experimental Sentinel-2 LAI estimation using parametric, non-parametric and physical retrieval methods—A comparison. ISPRS J. Photogramm. Remote Sens. 2015, 108, 260–272. [Google Scholar] [CrossRef]
- Blackburn, G.A. Wavelet decomposition of hyperspectral data: A novel approach to quantifying pigment concentrations in vegetation. Int. J. Remote Sens. 2007, 28, 2831–2855. [Google Scholar] [CrossRef]
- Geladi, P.; Kowalski, B.R. Partial least-squares regression: A tutorial. Anal. Chim. Acta 1986, 185, 1–17. [Google Scholar] [CrossRef]
- Kokaly, R.F.; Clark, R.N. Spectroscopic determination of leaf biochemistry using band-depth analysis of absorption features and stepwise multiple linear regression. Remote Sens. Environ. 1999, 67, 267–287. [Google Scholar] [CrossRef]
- Wold, S.; Esbensen, K.; Geladi, P. Principal component analysis. Chemom. Intell. Lab. Syst. 1987, 2, 37–52. [Google Scholar] [CrossRef]
- Breiman, L. Random forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef] [Green Version]
- Haykin, S. Neural Networks: A Comprehensive Foundation; Prentice Hall PTR: Hoboken, NJ, USA, 1994. [Google Scholar]
- Shawe-Taylor, J.; Cristianini, N. Kernel Methods for Pattern Analysis; Cambridge University Press: Cambridge, UK, 2004. [Google Scholar]
- Doughty, C.E.; Asner, G.P.; Martin, R.E. Predicting tropical plant physiology from leaf and canopy spectroscopy. Oecologia 2011, 165, 289–299. [Google Scholar] [CrossRef]
- Sun, J.; Shi, S.; Yang, J.; Du, L.; Gong, W.; Chen, B.; Song, S. Analyzing the performance of prospect model inversion based on different spectral information for leaf biochemical properties retrieval. ISPRS J. Photogramm. Remote Sens. 2018, 135, 74–83. [Google Scholar] [CrossRef]
- Jiang, J.; Comar, A.; Weiss, M.; Baret, F. Faspect: A model of leaf optical properties accounting for the differences between upper and lower faces. Remote Sens. Environ. 2021, 253, 112205. [Google Scholar] [CrossRef]
- Miraglio, T.; Adeline, K.; Huesca, M.; Ustin, S.; Briottet, X. Joint use of Prosail and dart for fast LUT building: Application to gap fraction and leaf biochemistry estimations over sparse oak stands. Remote Sens. 2020, 12, 2925. [Google Scholar] [CrossRef]
- Ali, A.M.; Darvishzadeh, R.; Skidmore, A.K. Retrieval of specific leaf area from Landsat-8 surface reflectance data using statistical and physical models. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2017, 10, 3529–3536. [Google Scholar] [CrossRef]
- Hosgood, B.; Jacquemoud, S.; Andreoli, G.; Verdebout, J.; Pedrini, G.; Schmuck, G. Leaf optical properties experiment 93 (LOPEX93). Rep. EUR 1995, 16095, 1–46. [Google Scholar]
- Hauser, L.T.; Féret, J.-B.; An Binh, N.; van der Windt, N.; Sil, Â.F.; Timmermans, J.; Soudzilovskaia, N.A.; van Bodegom, P.M. Towards scalable estimation of plant functional diversity from Sentinel-2: In-situ validation in a heterogeneous (semi-)natural landscape. Remote Sens. Environ. 2021, 262, 112505. [Google Scholar] [CrossRef]
- Faurtyot, T.; Baret, F. Vegetation water and dry matter contents estimated from top-of-the-atmosphere reflectance data: A simulation study. Remote Sens. Environ. 1997, 61, 34–45. [Google Scholar] [CrossRef]
- Darvishzadeh, R.; Skidmore, A.; Schlerf, M.; Atzberger, C.; Corsi, F.; Cho, M. LAI and chlorophyll estimation for a heterogeneous grassland using hyperspectral measurements. ISPRS J. Photogramm. Remote Sens. 2008, 63, 409–426. [Google Scholar] [CrossRef]
- Dormann, C.F.; Elith, J.; Bacher, S.; Buchmann, C.; Carl, G.; Carré, G.; Marquéz, J.R.G.; Gruber, B.; Lafourcade, B.; Leitão, P.J.; et al. Collinearity: A review of methods to deal with it and a simulation study evaluating their performance. Ecography 2013, 36, 27–46. [Google Scholar] [CrossRef]
- Grossman, Y.L.; Ustin, S.L.; Jacquemoud, S.; Sanderson, E.W.; Schmuck, G.; Verdebout, J. Critique of stepwise multiple linear regression for the extraction of leaf biochemistry information from leaf reflectance data. Remote Sens. Environ. 1996, 56, 182–193. [Google Scholar] [CrossRef]
- Streher, A.S.; Torres, R.d.S.; Morellato, L.P.C.; Silva, T.S.F. Accuracy and limitations for spectroscopic prediction of leaf traits in seasonally dry tropical environments. Remote Sens. Environ. 2020, 244, 111828. [Google Scholar] [CrossRef]
- Mutanga, O.; Skidmore, A.K. Narrow band vegetation indices overcome the saturation problem in biomass estimation. Int. J. Remote Sens. 2004, 25, 3999–4014. [Google Scholar] [CrossRef]
- Meerdink, S.K.; Roberts, D.A.; King, J.Y.; Roth, K.L.; Dennison, P.E.; Amaral, C.H.; Hook, S.J. Linking seasonal foliar traits to vswir-tir spectroscopy across california ecosystems. Remote Sens. Environ. 2016, 186, 322–338. [Google Scholar] [CrossRef]
- Rocha, A.D.; Groen, T.A.; Skidmore, A.K.; Darvishzadeh, R.; Willemen, L. The naïve overfitting index selection (NOIS): A new method to optimize model complexity for hyperspectral data. ISPRS J. Photogramm. Remote Sens. 2017, 133, 61–74. [Google Scholar] [CrossRef]
- Verrelst, J.; Rivera, J.P.; Leonenko, G.; Alonso, L.; Moreno, J. Optimizing LUT-based RTM inversion for semiautomatic mapping of crop biophysical parameters from Sentinel-2 and -3 data: Role of cost functions. IEEE Trans. Geosci. Remote Sens. 2014, 52, 257–269. [Google Scholar] [CrossRef]
- Nakaji, T.; Oguma, H.; Nakamura, M.; Kachina, P.; Asanok, L.; Marod, D.; Aiba, M.; Kurokawa, H.; Kosugi, Y.; Kassim, A.R.; et al. Estimation of six leaf traits of east asian forest tree species by leaf spectroscopy and partial least square regression. Remote Sens. Environ. 2019, 233, 111381. [Google Scholar] [CrossRef]
- Rocha, A.D.; Groen, T.A.; Skidmore, A.K.; Darvishzadeh, R.; Willemen, L. Machine learning using hyperspectral data inaccurately predicts plant traits under spatial dependency. Remote Sens. 2018, 10, 1263. [Google Scholar] [CrossRef] [Green Version]
- Combal, B.; Baret, F.; Weiss, M.; Trubuil, A.; Macé, D.; Pragnère, A.; Myneni, R.; Knyazikhin, Y.; Wang, L. Retrieval of canopy biophysical variables from bidirectional reflectance: Using prior information to solve the ill-posed inverse problem. Remote Sens. Environ. 2003, 84, 1–15. [Google Scholar] [CrossRef]
- Richter, K.; Atzberger, C.; Vuolo, F.; Weihs, P.; D’Urso, G. Experimental assessment of the Sentinel-2 band setting for RTM-based LAI retrieval of sugar beet and maize. Can. J. Remote Sens. 2009, 35, 230–247. [Google Scholar] [CrossRef]
- Spafford, L.; le Maire, G.; MacDougall, A.; de Boissieu, F.; Féret, J.-B. Spectral subdomains and prior estimation of leaf structure improves PROSPECT inversion on reflectance or transmittance alone. Remote Sens. Environ. 2021, 252, 112176. [Google Scholar] [CrossRef]
- Rivera-Caicedo, J.P.; Verrelst, J.; Muñoz-Marí, J.; Camps-Valls, G.; Moreno, J. Hyperspectral dimensionality reduction for biophysical variable statistical retrieval. ISPRS J. Photogramm. Remote Sens. 2017, 132, 88–101. [Google Scholar] [CrossRef]
- Skidmore, A.K.; Pettorelli, N.; Coops, N.C.; Geller, G.N.; Hansen, M.; Lucas, R.; Mücher, C.A.; O’Connor, B.; Paganini, M.; Pereira, H.M.; et al. Environmental science: Agree on biodiversity metrics to track from space. Nature 2015, 523, 403–405. [Google Scholar] [CrossRef] [Green Version]
- Pettorelli, N.; Wegmann, M.; Skidmore, A.; Mücher, S.; Dawson, T.P.; Fernandez, M.; Lucas, R.; Schaepman, M.E.; Wang, T.; O’Connor, B.; et al. Framing the concept of satellite remote sensing essential biodiversity variables: Challenges and future directions. Remote Sens. Ecol. Conserv. 2016, 2, 122–131. [Google Scholar] [CrossRef]
- Dawson, T.P.; Curran, P.J.; Plummer, S.E. Liberty—modeling the effects of leaf biochemical concentration on reflectance spectra. Remote Sens. Environ. 1998, 65, 50–60. [Google Scholar] [CrossRef]
- Verrelst, J.; Rivera, J.P.; Moreno, J.; Camps-Valls, G. Gaussian processes uncertainty estimates in experimental Sentinel-2 lai and leaf chlorophyll content retrieval. ISPRS J. Photogramm. Remote Sens. 2013, 86, 157–167. [Google Scholar] [CrossRef]
- Ali, A.M.; Darvishzadeh, R.; Skidmore, A.; Gara, T.W.; Heurich, M. Machine learning methods’ performance in radiative transfer model inversion to retrieve plant traits from Sentinel-2 data of a mixed mountain forest. Int. J. Digit. Earth 2021, 14, 106–120. [Google Scholar] [CrossRef]
- Ali, A.M.; Darvishzadeh, R.; Skidmore, A.; Gara, T.W.; O’Connor, B.; Roeoesli, C.; Heurich, M.; Paganini, M. Comparing methods for mapping canopy chlorophyll content in a mixed mountain forest using Sentinel-2 data. Int. J. Appl. Earth Obs. Geoinf. 2020, 87, 102037. [Google Scholar] [CrossRef]
- Ullah, S.; Skidmore, A.K.; Ramoelo, A.; Groen, T.A.; Naeem, M.; Ali, A. Retrieval of leaf water content spanning the visible to thermal infrared spectra. ISPRS J. Photogramm. Remote Sens. 2014, 93, 56–64. [Google Scholar] [CrossRef]
- Buitrago, M.F.; Groen, T.A.; Hecker, C.A.; Skidmore, A.K. Spectroscopic determination of leaf traits using infrared spectra. Int. J. Appl. Earth Obs. Geoinf. 2018, 69, 237–250. [Google Scholar] [CrossRef]
- Sibanda, M.; Mutanga, O.; Rouget, M. Comparing the spectral settings of the new generation broad and narrow band sensors in estimating biomass of native grasses grown under different management practices. GISci. Remote Sens. 2016, 53, 614–633. [Google Scholar] [CrossRef]
- Mitchell, J.J.; Shrestha, R.; Spaete, L.P.; Glenn, N.F. Combining airborne hyperspectral and lidar data across local sites for upscaling shrubland structural information: Lessons for hyspiri. Remote Sens. Environ. 2015, 167, 98–110. [Google Scholar] [CrossRef]
- Bell, T.W.; Cavanaugh, K.C.; Siegel, D.A. Remote monitoring of giant kelp biomass and physiological condition: An evaluation of the potential for the hyperspectral infrared imager (HYSPIRI) mission. Remote Sens. Environ. 2015, 167, 218–228. [Google Scholar] [CrossRef]
- Coble, A.P.; VanderWall, B.; Mau, A.; Cavaleri, M.A. How vertical patterns in leaf traits shift seasonally and the implications for modeling canopy photosynthesis in a temperate deciduous forest. Tree Physiol. 2016, 36, 1077–1091. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gara, T.; Darvishzadeh, R.; Skidmore, A.; Wang, T. Impact of vertical canopy position on leaf spectral properties and traits across multiple species. Remote Sens. 2018, 10, 346. [Google Scholar] [CrossRef] [Green Version]
- Kamoske, A.G.; Dahlin, K.M.; Serbin, S.P.; Stark, S.C. Leaf traits and canopy structure together explain canopy functional diversity: An airborne remote sensing approach. Ecol. Appl. 2020, 31, e02230. [Google Scholar] [CrossRef] [PubMed]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gara, T.W.; Rahimzadeh-Bajgiran, P.; Darvishzadeh, R. Forest Leaf Mass per Area (LMA) through the Eye of Optical Remote Sensing: A Review and Future Outlook. Remote Sens. 2021, 13, 3352. https://doi.org/10.3390/rs13173352
Gara TW, Rahimzadeh-Bajgiran P, Darvishzadeh R. Forest Leaf Mass per Area (LMA) through the Eye of Optical Remote Sensing: A Review and Future Outlook. Remote Sensing. 2021; 13(17):3352. https://doi.org/10.3390/rs13173352
Chicago/Turabian StyleGara, Tawanda W., Parinaz Rahimzadeh-Bajgiran, and Roshanak Darvishzadeh. 2021. "Forest Leaf Mass per Area (LMA) through the Eye of Optical Remote Sensing: A Review and Future Outlook" Remote Sensing 13, no. 17: 3352. https://doi.org/10.3390/rs13173352
APA StyleGara, T. W., Rahimzadeh-Bajgiran, P., & Darvishzadeh, R. (2021). Forest Leaf Mass per Area (LMA) through the Eye of Optical Remote Sensing: A Review and Future Outlook. Remote Sensing, 13(17), 3352. https://doi.org/10.3390/rs13173352