A Robust Prediction Model for Species Distribution Using Bagging Ensembles with Deep Neural Networks
Abstract
:1. Introduction
- We investigate several bias-minimizing methods, including selecting valuable environmental features and generating bootstraps from PA datasets. We use variance inflation factor (VIF) analysis to select suitable environmental features and use random sampling with replacement to generate multiple bootstraps.
- We predict species distribution using our ensemble DNNs trained with generated bootstraps, three voting methods, and repeated cross-validation to ensure reliable results. The generated models were compared to state-of-the-art practical SDMs using five evaluation metrics.
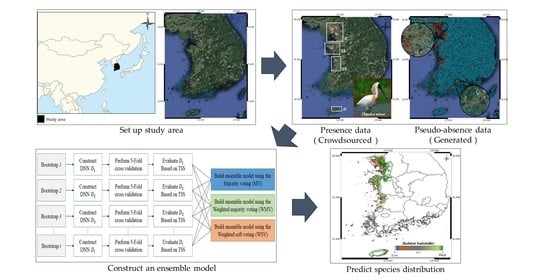
2. Methods
2.1. Dataset Construction
2.2. Pseudo-Absence and Bootstrap Generation
2.3. Ensemble Approach for Model Construction
2.4. Evaluation Approach
3. Experimental Results
3.1. Experimental Setting
3.2. Pseudo-Absence Generation Strategy Effects
3.3. SDM Stability for Unbalanced Datasets
3.4. Impact of the Ensemble Size
3.5. Case Study
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Tscharntke, T.; Clough, Y.; Wanger, T.C.; Jackson, L.; Motzke, I.; Perfecto, I.; Vandermeer, J.; Whitbread, A. Global food security, biodiversity conservation and the future of agricultural intensification. Biol. Conserv. 2012, 151, 53–59. [Google Scholar] [CrossRef]
- Potts, S.G.; Biesmeijer, J.C.; Kremen, C.; Neumann, P.; Schweiger, O.; Kunin, W.E. Global pollinator declines: Trends, impacts and drivers. Trends Ecol. Evol. 2010, 25, 345–353. [Google Scholar] [CrossRef] [PubMed]
- Collins, J.P.; Storfer, A. Global amphibian declines: Sorting the hypotheses. Divers. Distrib. 2003, 9, 89–98. [Google Scholar] [CrossRef]
- Wood, P.M. Biodiversity as the source of biological resources: A new look at biodiversity values. Environ. Values 1997, 6, 251–268. [Google Scholar] [CrossRef]
- Simpson, R.D.; Sedjo, R.A.; Reid, J.W. Valuing biodiversity for use in pharmaceutical research. J. Political Econ. 1996, 104, 163–185. [Google Scholar] [CrossRef]
- Butchart, S.H.; Walpole, M.; Collen, B.; Van Strien, A.; Scharlemann, J.P.; Almond, R.E.; Baillie, J.E.; Bomhard, B.; Brown, C.; Bruno, J.; et al. Global biodiversity: Indicators of recent declines. Science 2010, 328, 1164–1168. [Google Scholar] [CrossRef]
- Almond, R.; Grooten, M.; Peterson, T. Living Planet Report 2020—Bending the Curve of Biodiversity Loss; World Wildlife Fund: Gland, Switzerland, 2020. [Google Scholar]
- Wilcove, D.S.; Rothstein, D.; Dubow, J.; Phillips, A.; Losos, E. Quantifying threats to imperiled species in the United States. BioScience 1998, 48, 607–615. [Google Scholar] [CrossRef] [Green Version]
- Langpap, C.; Kerkvliet, J. Endangered species conservation on private land: Assessing the effectiveness of habitat conservation plans. J. Environ. Econ. Manag. 2012, 64, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Bonnie, R. Endangered species mitigation banking: Promoting recovery through habitat conservation planning under the Endangered Species Act. Sci. Total Environ. 1999, 240, 11–19. [Google Scholar] [CrossRef]
- Elith, J. Quantitative Methods for Modeling Species Habitat: Comparative Performance and an Application to Australian Plants; Springer: New York, NY, USA, 2006; pp. 39–58. [Google Scholar]
- Braunisch, V.; Suchant, R. A model for evaluating the ‘habitat potential’ of a landscape for capercaillie Tetrao urogallus: A tool for conservation planning. Wildl. Biol. 2007, 13, 21–33. [Google Scholar] [CrossRef] [Green Version]
- Wu, X.B.; Smeins, F.E. Multiple-scale habitat modeling approach for rare plant conservation. Landsc. Urban Plan. 2000, 51, 11–28. [Google Scholar] [CrossRef]
- Poulos, H.M.; Chernoff, B.; Fuller, P.L.; Butman, D. Ensemble forecasting of potential habitat for three invasive fishes. Aquat. Invasions 2012, 7, 59–72. [Google Scholar] [CrossRef]
- Brown, J.L. SDMtoolbox: A python-based GIS toolkit for landscape genetic, biogeographic and species distribution model analyses. Methods Ecol. Evol. 2014, 5, 694–700. [Google Scholar] [CrossRef]
- Václavík, T.; Meentemeyer, R.K. Equilibrium or not? Modelling potential distribution of invasive species in different stages of invasion. Divers. Distrib. 2011, 18, 73–83. [Google Scholar] [CrossRef]
- Robinson, T.P.; van Klinken, R.D.; Metternicht, G. Comparison of alternative strategies for invasive species distribution modeling. Ecol. Model. 2010, 221, 2261–2269. [Google Scholar] [CrossRef] [Green Version]
- Raes, N.; ter Steege, H. A null-model for significance testing of presence-only species distribution models. Ecography 2007, 30, 727–736. [Google Scholar] [CrossRef]
- Zaniewski, A.E.; Lehmann, A.; Overton, J.M. Predicting species spatial distributions using presence-only data: A case study of native New Zealand ferns. Ecol. Model. 2002, 157, 261–280. [Google Scholar] [CrossRef]
- Rebelo, H.; Jones, G. Ground validation of presence-only modelling with rare species: A case study on barbastelles Barbastella barbastellus (Chiroptera: Vespertilionidae). J. Appl. Ecol. 2010, 47, 410–420. [Google Scholar] [CrossRef]
- Aarts, G.; Fieberg, J.; Matthiopoulos, J. Comparative interpretation of count, presence-absence and point methods for species distribution models. Methods Ecol. Evol. 2012, 3, 177–187. [Google Scholar] [CrossRef]
- Elith, J.; Graham, C.H. Do they? How do they? WHY do they differ? On finding reasons for differing performances of species distribution models. Ecography 2009, 32, 66–77. [Google Scholar] [CrossRef]
- Manel, S.; Williams, H.C.; Ormerod, S.J. Evaluating presence-absence models in ecology: The need to account for prevalence. J. Appl. Ecol. 2001, 38, 921–931. [Google Scholar] [CrossRef]
- Duan, R.-Y.; Kong, X.-Q.; Huang, M.-Y.; Fan, W.-Y.; Wang, Z.-G. The predictive performance and stability of six species distribution models. PLoS ONE 2014, 9, e112764. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Munguía, M.; Rahbek, C.; Rangel, T.F.; Diniz-Filho, J.A.F.; Araújo, M.B. Equilibrium of global amphibian species distributions with climate. PLoS ONE 2012, 7, e34420. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hampe, A. Bioclimate envelope models: What they detect and what they hide. Glob. Ecol. Biogeogr. 2004, 13, 469–471. [Google Scholar] [CrossRef]
- Sillero, N. What does ecological modelling model? A proposed classification of ecological niche models based on their under-lying methods. Ecol. Model. 2011, 222, 1343–1346. [Google Scholar] [CrossRef]
- Barve, N.; Barve, V.; Jiménez-Valverde, A.; Lira-Noriega, A.; Maher, S.P.; Peterson, A.T.; Soberón, J.; Villalobos, F. The crucial role of the accessible area in ecological niche modeling and species distribution modeling. Ecol. Model. 2011, 222, 1810–1819. [Google Scholar] [CrossRef]
- Phillips, N.D.; Reid, N.; Thys, T.; Harrod, C.; Payne, N.L.; Morgan, C.A.; White, H.J.; Porter, S.; Houghton, J.D. Applying species distribution modelling to a data poor, pelagic fish complex: The ocean sunfishes. J. Biogeogr. 2017, 44, 2176–2187. [Google Scholar] [CrossRef]
- Reiss, H.; Cunze, S.; König, K.; Neumann, H.; Kröncke, I. Species distribution modelling of marine benthos: A North Sea case study. Mar. Ecol. Prog. Ser. 2011, 442, 71–86. [Google Scholar] [CrossRef] [Green Version]
- Guisan, A.; Thuiller, W. Predicting species distribution: Offering more than simple habitat models. Ecol. Lett. 2005, 8, 993–1009. [Google Scholar] [CrossRef]
- Thomaes, A.; Kervyn, T.; Maes, D. Applying species distribution modelling for the conservation of the threatened saproxylic Stag Beetle (Lucanus cervus). Biol. Conserv. 2008, 141, 1400–1410. [Google Scholar] [CrossRef]
- De’ath, G.; Fabricius, K.E. Classification and regression trees: A powerful yet simple technique for ecological data analysis. Ecology 2000, 81, 3178–3192. [Google Scholar] [CrossRef]
- De’ath, G. Boosted trees for ecological modeling and prediction. Ecology 2007, 88, 243–251. [Google Scholar] [CrossRef]
- D’Heygere, T.; Goethals, P.L.; De Pauw, N. Genetic algorithms for optimisation of predictive ecosystems models based on decision trees and neural networks. Ecol. Model. 2006, 195, 20–29. [Google Scholar] [CrossRef]
- Bird, T.J.; Bates, A.E.; Lefcheck, J.S.; Hill, N.A.; Thomson, R.J.; Edgar, G.J.; Stuart-Smith, R.D.; Wotherspoon, S.; Krkosek, M.; Stuart-Smith, J.F.; et al. Statistical solutions for error and bias in global citizen science datasets. Biol. Conserv. 2014, 173, 144–154. [Google Scholar] [CrossRef] [Green Version]
- Geldmann, J.; Heilmann-Clausen, J.; Holm, T.E.; Levinsky, I.; Markussen, B.; Olsen, K.; Rahbek, C.; Tøttrup, A.P. What determines spatial bias in citizen science? Exploring four recording schemes with different proficiency requirements. Divers. Distrib. 2016, 22, 1139–1149. [Google Scholar] [CrossRef]
- Rademaker, M.; Hogeweg, L.; Vos, R. Modelling the niches of wild and domesticated Ungulate species using deep learning. bioRxiv 2019, 744441. [Google Scholar] [CrossRef]
- Botella, C.; Joly, A.; Bonnet, P.; Monestiez, P.; Munoz, F. A Deep Learning Approach to Species Distribution Modelling; Springer: Cham, Switzerland, 2018; pp. 169–199. [Google Scholar]
- Benkendorf, D.J.; Hawkins, C.P. Effects of sample size and network depth on a deep learning approach to species distribution modeling. Ecol. Inform. 2020, 60, 101137. [Google Scholar] [CrossRef]
- GBIF Homepage. Available online: https://www.gbif.org (accessed on 22 November 2020).
- VertNet Homepage. Available online: http://vertnet.org (accessed on 22 November 2020).
- BISON Homepage. Available online: https://bison.usgs.gov (accessed on 22 November 2020).
- Naturing Homepage. Available online: https://www.naturing.net (accessed on 22 November 2020).
- GBIF.org. GBIF Occurrence Download. Available online: https://bit.ly/3a0rwZ2 (accessed on 12 April 2021). [CrossRef]
- GBIF.org. GBIF Occurrence Download. Available online: https://bit.ly/3sjPW6l (accessed on 12 April 2021). [CrossRef]
- GBIF.org. GBIF Occurrence Download. Available online: https://bit.ly/3s8726R (accessed on 12 April 2021). [CrossRef]
- GBIF.org. GBIF Occurrence Download. Available online: https://bit.ly/2PV798Q (accessed on 12 April 2021). [CrossRef]
- GBIF.org. GBIF Occurrence Download. Available online: https://bit.ly/3wOD6jO (accessed on 12 April 2021). [CrossRef]
- Hernandez, P.A.; Graham, C.H.; Master, L.L.; Albert, D.L. The effect of sample size and species characteristics on performance of different species distribution modeling methods. Ecography 2006, 29, 773–785. [Google Scholar] [CrossRef]
- Stockwell, D.R.; Peterson, A.T. Effects of sample size on accuracy of species distribution models. Ecol. Model. 2002, 148, 1–13. [Google Scholar] [CrossRef]
- Aiello-Lammens, M.E.; Boria, R.A.; Radosavljevic, A.; Vilela, B.; Anderson, R.P. spThin: An R package for spatial thinning of species occurrence records for use in ecological niche models. Ecography 2015, 38, 541–545. [Google Scholar] [CrossRef]
- Fick, S.E.; Hijmans, R.J. WorldClim 2: New 1-km spatial resolution climate surfaces for global land areas. Int. J. Climatol. 2017, 37, 4302–4315. [Google Scholar] [CrossRef]
- Arino, O.; Perez, J.R.; Kalogirou, V.; Bontemps, S.; Defourny, P.; van Bogaert, E. Global Land Cover Map for 2009 (GlobCover 2009); European Space Agency (ESA); Université Catholique de Louvain (UCL): Frascati, Italy, 2012. [Google Scholar]
- Naimi, B.; Hamm, N.A.; Groen, T.A.; Skidmore, A.K.; Toxopeus, A.G. Where is positional uncertainty a problem for species distribution modelling? Ecography 2013, 37, 191–203. [Google Scholar] [CrossRef]
- Barbet-Massin, M.; Jiguet, F.; Albert, C.H.; Thuiller, W. Selecting pseudo-absences for species distribution models: How, where and how many? Methods Ecol. Evol. 2012, 3, 327–338. [Google Scholar] [CrossRef]
- Iturbide, M.; Bedia, J.; Herrera, S.; del Hierro, O.; Pinto, M.; Gutiérrez, J.M. A framework for species distribution modelling with improved pseudo-absence generation. Ecol. Model. 2015, 312, 166–174. [Google Scholar] [CrossRef] [Green Version]
- Chefaoui, R.M.; Lobo, J.M. Assessing the effects of pseudo-absences on predictive distribution model performance. Ecol. Model. 2008, 210, 478–486. [Google Scholar] [CrossRef]
- Iturbide, M.; Bedia, J.; Gutiérrez, J. Tackling Uncertainties of Species Distribution Model Projections with Package mopa. R J. 2018, 10, 122–139. [Google Scholar] [CrossRef] [Green Version]
- Chernick, M. Bootstrap Methods: A Guide for Researchers and Practitioners; Wiley: New York, NY, USA, 2007. [Google Scholar] [CrossRef]
- Jung, S.; Moon, J.; Park, S.; Rho, S.; Baik, S.W.; Hwang, E. Bagging ensemble of multilayer perceptrons for missing electricity consumption data imputation. Sensors 2020, 20, 1772. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Canty, A.J. Resampling Methods in R: The Boot Package. The Newsletter of the R Project, December 2002, Volume 2/3. Available online: http://cran.fhcrc.org/doc/Rnews/Rnews_2002-3.pdf (accessed on 12 April 2021).
- Rew, J.; Cho, Y.; Moon, J.; Hwang, E. Habitat Suitability Estimation Using a Two-Stage Ensemble Approach. Remote Sens. 2020, 12, 1475. [Google Scholar] [CrossRef]
- Thuiller, W.; Lafourcade, B.; Engler, R.; Araújo, M.B. BIOMOD—A platform for ensemble forecasting of species distributions. Ecography 2009, 32, 369–373. [Google Scholar] [CrossRef]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V. Scikit-learn: Machine learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Kang, J.-H.; Kim, I.K.; Lee, K.-S.; Lee, H.; Rhim, S.-J. Distribution, breeding status, and conservation of the black-faced spoonbill (Platalea minor) in South Korea. For. Sci. Technol. 2016, 12, 162–166. [Google Scholar] [CrossRef] [Green Version]
- Kang, J.-H.; Kim, I.K.; Lee, K.-S.; Kwon, I.-K.; Lee, H.; Rhim, S.-J. Home range and movement of juvenile black-faced spoonbill Platalea minor in South Korea. J. Ecol. Environ. 2017, 41, 1–5. [Google Scholar] [CrossRef] [Green Version]
- Engler, R.; Guisan, A.; Rechsteiner, L. An improved approach for predicting the distribution of rare and endangered species from occurrence and pseudo-absence data. J. Appl. Ecol. 2004, 41, 263–274. [Google Scholar] [CrossRef]
















| Scientific Name | Sample Image | IUCN Red List Grade 1 | Total Observations in South Korea | Total Presences after Spatial Bias Removal | Suitable Habitats |
|---|---|---|---|---|---|
| Hynobius leechii |  | LC | 1432 | 1024 | - Forest - Wetlands - Freshwater marches |
| Cyanopica cyanus |  | LC | 3412 | 2666 | - Forest, - Moist lowland |
| Platalea minor |  | EN | 1327 | 1078 | - Marine intertidal - Marine coastal /supratidal (sea cliffs and rocky) |
| Hypsipetes amaurotis |  | LC | 8406 | 6401 | - Subtropical /tropical forest - Moist lowland |
| Hyla japonica |  | LC | 3125 | 2338 | - Forest /grassland - Scrubland - Wetland - Arable land - Pastureland - Rural gardens - Urban areas |
| Variable Name | Description | Data Type | Spatial Resolution |
|---|---|---|---|
| Climate_01 | Annual mean temperature | Continuous | 30 s |
| Climate_02 | Mean diurnal range | Continuous | 30 s |
| Climate_03 | Isothermality | Continuous | 30 s |
| Climate_04 | Temperature seasonality | Continuous | 30 s |
| Climate_05 | Max temperature of warmest month | Continuous | 30 s |
| Climate_06 | Min temperature of coldest month | Continuous | 30 s |
| Climate_07 | Temperature annual range | Continuous | 30 s |
| Climate_08 | Mean temperature of wettest quarter | Continuous | 30 s |
| Climate_09 | Mean temperature of driest quarter | Continuous | 30 s |
| Climate_10 | Mean temperature of warmest quarter | Continuous | 30 s |
| Climate_11 | Mean temperature of coldest quarter | Continuous | 30 s |
| Climate_12 | Annual precipitation | Continuous | 30 s |
| Climate_13 | Precipitation of wettest month | Continuous | 30 s |
| Climate_14 | Precipitation of driest month | Continuous | 30 s |
| Climate_15 | Precipitation seasonality | Continuous | 30 s |
| Climate_16 | Precipitation of wettest quarter | Continuous | 30 s |
| Climate_17 | Precipitation of driest quarter | Continuous | 30 s |
| Climate_18 | Precipitation of warmest quarter | Continuous | 30 s |
| Climate_19 | Precipitation of coldest quarter | Continuous | 30 s |
| GlobCover_01 | Rainfed croplands | Boolean | 300 m |
| GlobCover_02 | Mosaic cropland (50–70%)/vegetation (20–50%) | Boolean | 300 m |
| GlobCover_03 | Mosaic vegetation (50–70%)/cropland (20–50%) | Boolean | 300 m |
| GlobCover_04 | Closed (>40%) broadleaved deciduous forest (>5 m) | Boolean | 300 m |
| GlobCover_05 | Closed (>40%) needle leaved evergreen forest (>5 m) | Boolean | 300 m |
| GlobCover_06 | Open (15–40%) needle leaved deciduous or evergreen forest (>5 m) | Boolean | 300 m |
| GlobCover_07 | Closed to open (>15%) mixed broadleaved/needle leaved forest (>5 m) | Boolean | 300 m |
| GlobCover_08 | Mosaic forest or shrubland (50–70%)/grassland (20–50%) | Boolean | 300 m |
| GlobCover_09 | Mosaic grassland (50%–70%)/forest or shrubland (20%–50%) | Boolean | 300 m |
| GlobCover_10 | Closed to open (>15%) herbaceous vegetation | Boolean | 300 m |
| GlobCover_11 | Sparse (<15%) vegetation | Boolean | 300 m |
| GlobCover_12 | Artificial surfaces and associated areas (urban areas >50%) | Boolean | 300 m |
| GlobCover_13 | Bare areas | Boolean | 300 m |
| GlobCover_14 | Water bodies | Boolean | 300 m |
| Predicted Present | Predicted Absent | |
|---|---|---|
| Actually present | True positive | False negative |
| Actually absent | False positive | True negative |
| AUC | K | TSS | |
|---|---|---|---|
| Excellent | |||
| Good | |||
| Fair | |||
| Poor or no predictive ability |
| Prediction Model | Training Strategies and Selected Parameters | Modeling Software |
|---|---|---|
| GLM | Quadratic regression Akaike information criterion for environmental layer selection | BIOMOD2 (R) |
| GBM | Bernoulli distribution, 2500 trees, 7 depths, 5 terminal nodes, 0.001 learning rate | BIOMOD2 (R) |
| CTA | Categorical classification, default tree parameter (auto-optimized by BIOMOD2) | BIOMOD2 (R) |
| SNN | Single hidden layer, auto-optimized neuron size, 200 iterations | BIOMOD2 (R) |
| FDA | MARSs method | BIOMOD2 (R) |
| MARS | Simple piecewise linear, 0.001 threshold, backward pruning | BIOMOD2 (R) |
| RF | Maximum 500 trees, default number of variables at each split (auto-optimized by BIOMOD2), 5 nodes | BIOMOD2 (R) |
| SRE | 0.025 quantile for environmental variable selection | BIOMOD2 (R) |
| MAXENT | Maximum 200 iterations, linear and quadratic variables, default parameters for threshold and hinge (auto-optimized by BIOMOD2) | BIOMOD2 (R) |
| DNN | 4 hidden layers, using dropout, 10,000 iterations with early stopping, ReLU, ADAM optimizer | Scikit-learn (Python) |
| MV-EDNN | 10 bootstraps, 5-fold cross validation of each bootstrap | Scikit-learn (Python) |
| WMV-EDNN | 10 bootstraps, weights using TSS evaluation, 5-fold cross validation of each bootstrap | Scikit-learn (Python) |
| WSV-EDNN | 10 bootstraps, weights using TSS evaluation, 5-fold cross validation of each bootstrap | Scikit-learn (Python) |
| SDM Type | Mean Evaluation Metric | ||||
|---|---|---|---|---|---|
| Sensitivity | Specificity | AUC | Κ | TSS | |
| GLM | 0.346 | 0.988 | 0.850 | 0.335 | 0.335 |
| SNN | 0.404 | 0.991 | 0.836 | 0.395 | 0.396 |
| MARS | 0.364 | 0.995 | 0.871 | 0.360 | 0.360 |
| RF | 0.778 | 0.993 | 0.890 | 0.771 | 0.771 |
| GBM | 0.482 | 0.996 | 0.878 | 0.479 | 0.479 |
| MAXENT | 0.517 | 0.926 | 0.822 | 0.444 | 0.444 |
| SRE | 0.705 | 0.727 * | 0.716 * | 0.432 | 0.433 |
| CTA | 0.630 | 0.989 | 0.878 | 0.620 | 0.620 |
| FDA | 0.375 | 0.988 | 0.835 | 0.363 | 0.363 |
| DNN | 0.589 | 0.676 * | 0.653 * | 0.235 | 0.266 |
| MV-EDNN | 0.896 | 0.900 | 0.898 | 0.796 | 0.797 |
| WMV-EDNN | 0.892 | 0.906 | 0.899 | 0.798 | 0.799 |
| WSV-EDNN | 0.889 | 0.907 | 0.898 | 0.795 | 0.796 |
| SDM Type | Mean Evaluation Metric | ||||
|---|---|---|---|---|---|
| Sensitivity | Specificity | AUC | Κ | TSS | |
| GLM | 0.589 | 0.987 | 0.936 | 0.576 | 0.576 |
| SNN | 0.645 | 0.992 | 0.909 | 0.638 | 0.638 |
| MARS | 0.584 | 0.986 | 0.938 | 0.570 | 0.570 |
| RF | 0.933 | 0.993 | 0.959 | 0.930 | 0.932 |
| GBM | 0.610 | 0.991 | 0.950 | 0.604 | 0.604 |
| MAXENT | 0.591 | 0.994 | 0.845 | 0.585 | 0.585 |
| SRE | 0.591 | 0.991 | 0.716 * | 0.585 | 0.585 |
| CTA | 0.710 | 0.721 * | 0.938 | 0.431 | 0.431 |
| FDA | 0.792 | 0.990 | 0.919 | 0.786 | 0.786 |
| DNN | 0.570 | 0.982 | 0.849 | 0.552 | 0.552 |
| MV-EDNN | 0.756 | 0.937 | 0.979 | 0.627 | 0.693 |
| WMV-EDNN | 0.977 | 0.954 | 0.979 | 0.931 | 0.931 |
| WSV-EDNN | 0.975 | 0.957 | 0.979 | 0.932 | 0.933 |
| SDM Type | Mean Evaluation Metric | ||||
|---|---|---|---|---|---|
| Sensitivity | Specificity | AUC | Κ | TSS | |
| GLM | 0.633 | 0.911 | 0.927 | 0.674 | 0.544 |
| SNN | 0.681 | 0.921 | 0.922 | 0.755 | 0.602 |
| MARS | 0.638 | 0.933 | 0.937 | 0.702 | 0.571 |
| RF | 0.932 | 0.97 | 0.951 | 0.94 | 0.902 |
| GBM | 0.691 | 0.925 | 0.952 | 0.758 | 0.616 |
| MAXENT | 0.655 | 0.927 | 0.854 | 0.693 | 0.582 |
| SRE | 0.72 | 0.788 * | 0.776 * | 0.614 | 0.508 |
| CTA | 0.733 | 0.971 | 0.943 | 0.68 | 0.704 |
| FDA | 0.714 | 0.924 | 0.915 | 0.688 | 0.638 |
| DNN | 0.688 | 0.918 | 0.88 | 0.642 | 0.606 |
| MV-EDNN | 0.952 | 0.954 | 0.965 | 0.949 | 0.906 |
| WMV-EDNN | 0.977 | 0.971 | 0.966 | 0.958 | 0.948 |
| WSV-EDNN | 0.985 | 0.973 | 0.964 | 0.957 | 0.958 |
| SDM Type | Sensitivity | Specificity | AUC | Κ | TSS | |
|---|---|---|---|---|---|---|
| 0.5 | GLM | 0.633 | 0.911 | 0.927 | 0.674 | 0.544 |
| SNN | 0.681 | 0.921 | 0.922 | 0.755 | 0.602 | |
| MARS | 0.638 | 0.933 | 0.937 | 0.702 | 0.571 | |
| RF | 0.932 | 0.970 | 0.951 | 0.940 | 0.902 | |
| GBM | 0.691 | 0.925 | 0.952 | 0.758 | 0.616 | |
| MAXENT | 0.655 | 0.927 | 0.854 | 0.693 | 0.582 | |
| SRE | 0.720 | 0.788 | 0.776 | 0.614 | 0.508 | |
| CTA | 0.733 | 0.971 | 0.943 | 0.680 | 0.704 | |
| FDA | 0.714 | 0.924 | 0.915 | 0.688 | 0.638 | |
| DNN | 0.688 | 0.918 | 0.88 | 0.642 | 0.606 | |
| MV-EDNNs | 0.952 | 0.954 | 0.965 | 0.949 | 0.906 | |
| WMV-EDNNs | 0.977 | 0.971 | 0.966 | 0.958 | 0.948 | |
| WSV-EDNNs | 0.985 | 0.973 | 0.964 | 0.957 | 0.958 | |
| 0.4 | GLM | 0.625 | 0.932 | 0.92 | 0.674 | 0.557 |
| SNN | 0.669 | 0.932 | 0.915 | 0.757 | 0.601 | |
| MARS | 0.625 | 0.942 | 0.931 | 0.708 | 0.567 | |
| RF | 0.930 | 0.972 | 0.955 | 0.948 | 0.902 | |
| GBM | 0.668 | 0.937 | 0.945 | 0.751 | 0.605 | |
| MAXENT | 0.628 | 0.942 | 0.850 | 0.694 | 0.57 | |
| SRE | 0.714 | 0.801 | 0.779 | 0.507 | 0.515 | |
| CTA | 0.722 | 0.973 | 0.949 | 0.675 | 0.695 | |
| FDA | 0.645 | 0.942 | 0.910 | 0.692 | 0.587 | |
| DNN | 0.662 | 0.922 | 0.878 | 0.614 | 0.584 | |
| MV-EDNNs | 0.944 | 0.958 | 0.961 | 0.945 | 0.902 | |
| WMV-EDNNs | 0.965 | 0.978 | 0.972 | 0.952 | 0.943 | |
| WSV-EDNNs | 0.962 | 0.975 | 0.971 | 0.957 | 0.937 | |
| 0.33 | GLM | 0.589 | 0.965 | 0.92 | 0.648 | 0.554 |
| SNN | 0.621 | 0.965 | 0.916 | 0.723 | 0.586 | |
| MARS | 0.615 | 0.952 | 0.934 | 0.693 | 0.567 | |
| RF | 0.933 | 0.958 | 0.954 | 0.941 | 0.921 | |
| GBM | 0.632 | 0.942 | 0.934 | 0.734 | 0.574 | |
| MAXENT | 0.607 | 0.962 | 0.849 | 0.668 | 0.569 | |
| SRE | 0.711 | 0.81 | 0.734 | 0.528 | 0.521 | |
| CTA | 0.727 | 0.981 | 0.913 | 0.668 | 0.708 | |
| FDA | 0.635 | 0.952 | 0.911 | 0.669 | 0.587 | |
| DNN | 0.626 | 0.957 | 0.876 | 0.602 | 0.583 | |
| MV-EDNNs | 0.942 | 0.970 | 0.963 | 0.936 | 0.912 | |
| WMV-EDNNs | 0.947 | 0.986 | 0.963 | 0.946 | 0.932 | |
| WSV-EDNNs | 0.945 | 0.985 | 0.963 | 0.949 | 0.930 | |
| 0.25 | GLM | 0.553 | 0.972 | 0.921 | 0.632 | 0.525 |
| SNN | 0.608 | 0.973 | 0.916 | 0.711 | 0.581 | |
| MARS | 0.609 | 0.965 | 0.934 | 0.666 | 0.574 | |
| RF | 0.926 | 0.985 | 0.950 | 0.942 | 0.911 | |
| GBM | 0.620 | 0.950 | 0.935 | 0.716 | 0.570 | |
| MAXENT | 0.585 | 0.970 | 0.855 | 0.668 | 0.555 | |
| SRE | 0.690 | 0.822 | 0.778 | 0.557 | 0.512 | |
| CTA | 0.710 | 0.991 | 0.938 | 0.651 | 0.701 | |
| FDA | 0.630 | 0.977 | 0.915 | 0.651 | 0.607 | |
| DNN | 0.621 | 0.975 | 0.864 | 0.598 | 0.595 | |
| MV-EDNNs | 0.925 | 0.982 | 0.963 | 0.921 | 0.907 | |
| WMV-EDNNs | 0.935 | 0.989 | 0.964 | 0.950 | 0.924 | |
| WSV-EDNNs | 0.929 | 0.989 | 0.964 | 0.940 | 0.918 | |
| 0.20 | GLM | 0.521 | 0.985 | 0.919 | 0.589 | 0.506 |
| SNN | 0.601 | 0.991 | 0.924 | 0.673 | 0.592 | |
| MARS | 0.568 | 0.990 | 0.932 | 0.618 | 0.558 | |
| RF | 0.925 | 0.998 | 0.963 | 0.937 | 0.922 | |
| GBM | 0.601 | 0.994 | 0.934 | 0.676 | 0.595 | |
| MAXENT | 0.535 | 0.988 | 0.845 | 0.614 | 0.523 | |
| SRE | 0.684 | 0.835 | 0.777 | 0.477 | 0.519 | |
| CTA | 0.625 | 0.995 | 0.947 | 0.651 | 0.620 | |
| FDA | 0.605 | 0.986 | 0.914 | 0.612 | 0.591 | |
| DNN | 0.584 | 0.986 | 0.852 | 0.527 | 0.57 | |
| MV-EDNNs | 0.910 | 0.992 | 0.968 | 0.918 | 0.902 | |
| WMV-EDNNs | 0.911 | 0.992 | 0.969 | 0.938 | 0.903 | |
| WSV-EDNNs | 0.913 | 0.992 | 0.968 | 0.936 | 0.905 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rew, J.; Cho, Y.; Hwang, E. A Robust Prediction Model for Species Distribution Using Bagging Ensembles with Deep Neural Networks. Remote Sens. 2021, 13, 1495. https://doi.org/10.3390/rs13081495
Rew J, Cho Y, Hwang E. A Robust Prediction Model for Species Distribution Using Bagging Ensembles with Deep Neural Networks. Remote Sensing. 2021; 13(8):1495. https://doi.org/10.3390/rs13081495
Chicago/Turabian StyleRew, Jehyeok, Yongjang Cho, and Eenjun Hwang. 2021. "A Robust Prediction Model for Species Distribution Using Bagging Ensembles with Deep Neural Networks" Remote Sensing 13, no. 8: 1495. https://doi.org/10.3390/rs13081495
APA StyleRew, J., Cho, Y., & Hwang, E. (2021). A Robust Prediction Model for Species Distribution Using Bagging Ensembles with Deep Neural Networks. Remote Sensing, 13(8), 1495. https://doi.org/10.3390/rs13081495