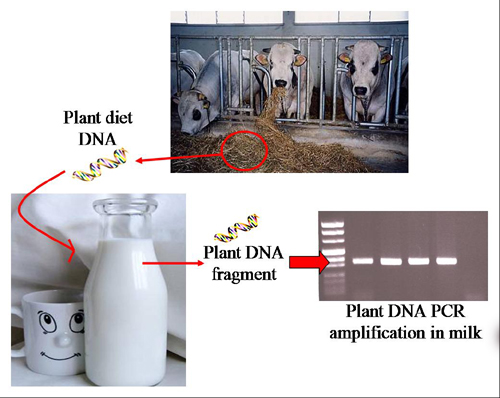
Traceability of Plant Diet Contents in Raw Cow Milk Samples
Abstract
:1. Introduction
2. Results and Discussion
2.1. Optimization of Plant DNA Extraction in Milk
| DNA extraction method used | Starting material | DNA quality A260/A280 | DNA yield/mL of milk |
|---|---|---|---|
| CTAB-NaCl buffer protocol | 20 mL | 0.98 | 190 μg |
| CTAB/phenol:chloroform protocol | 20 mL | 1.5 | 950 μg |
| Hexane protocol skim fraction | 17 mL | 1.4 | 460 μg |
| Hexane protocol fat fraction | 3 mL | 1.3 | 620 μg |

2.2. Feed-Derived Plant DNA Detection in Different Milk Samples

2.3. Reproducibility of Plant DNA Detection in Cow Milk Samples

2.4. Sequencing of Plant DNA Fragments Amplified in Cow Milk Samples

3. Experimental Section
3.1. Total Milk DNA Extraction
3.2. PCR Methodology and Primer Design
3.3. Sequence Analyses
4. Conclusions
Acknowledgements
References and Notes
- Coff, C.; Korthals, M.; Barling, D. Chapter 1: Ethical Traceability and Informed Food Choice. In Ethical Traceability and Communicating Food; Coff, C., Barling, D., Korthals, M., Nielsen, T., Eds.; Springer: Dordrecht, The Netherlands, 2008; Volume 15, pp. 1–18. [Google Scholar]
- Mafra, I.; Ferreira, I.M.P.L.V.O.; Oliveira, M.B.P.P. Food authentication by PCR-based methods. Eur. Food Res. Technol. 2008, 227, 649–665. [Google Scholar]
- Lopez-Calleja, I.; Gonzalez, I.; Fajardo, V.; Rodriguez, M.A.; Hernandez, P.E.; Garcia, T.; Martin, R. Rapid detection of cows’ milk in sheeps’ and goats’ milk by a species-specific polymerase chain reaction technique. J. Dairy Sci. 2004, 87, 2839–2845. [Google Scholar]
- Lanzilao, I.; Burgalassi, F.; Fancelli, S.; Settimelli, M.; Fani, R. Polymerase chain reaction-restriction fragment length polymorphism analysis of mitochondrial cytb gene from species of dairy interest. J. AOAC Int. 2005, 88, 128–135. [Google Scholar] [PubMed]
- Breviario, D.; Morello, L.; Manca, A.; Gianì, S. Chapter 2: The Importance of Being an Intron, by Wild Type Tubulin Genes. In The Plant Cytoskeleton: a Key Tool for Agro-Biotechnology; Blume, Y.B., Baird, W.V., Yemets, A.I., Breviario, D., Eds.; Springer: Dordrecht, The Netherlands, 2008; pp. 199–218. [Google Scholar]
- Schubbert, R.; Renz, D.; Schmitz, B.; Doerfler, W. Foreign (M13) DNA ingested by mice reaches peripheral leukocytes, spleen, and liver via the intestinal wall mucosa and can be covalently linked to mouse DNA. Proc. Natl. Acad. Sci. USA 1997, 94, 961–966. [Google Scholar]
- Schubbert, R.; Hohlweg, U.; Renz, D.; Doerfler, W. On the fate of orally ingested foreign DNA in mice: chromosomal association and placental transmission to the foetus. Mol. Gen. Genet. 1998, 259, 569–576. [Google Scholar]
- Tony, M.A.; Butschke, A.; Broll, H.; Grohmann, L.; Zagon, J.; Halle, I.; Dänicke, S.; Schauzu, M.; Flachowsky, G. Safety assessment of Bt 176 maize in broiler nutrition: Degradation of maize-DNA and its metabolic fate. Arch. Anim. Nutr. 2003, 57, 235–252. [Google Scholar]
- Einspanier, R.; Klotz, A.; Kraft, J.; Aulrich, K.; Poser, R.; Schwägele, F.; Jahreis, G.; Flachowsky, G. The fate of forage plant DNA in farm animals: a collaborative case-study investigating cattle and chicken fed recombinant plant material. Eur. Food Res. Tech. 2001, 212, 129–134. [Google Scholar] [CrossRef]
- Reuter, T.; Aulrich, K.; Berk, A.; Flachowsky, G. Investigations on genetically modified corn (Bt-corn) in pig nutrition: Chemical composition and nutritional evaluation. Arch. Anim. Nutr. 2002, 56, 23–31. [Google Scholar]
- Phipps, R.H.; Deaville, E.R.; Maddison, B.C. Detection of transgenic and endogenous plant DNA in rumen fluid, duodenal digesta, milk, blood, and feces of lactating dairy cows. J. Dairy Sci. 2003, 86, 4070–4078. [Google Scholar] [CrossRef] [PubMed]
- Nemeth, A.; Wurz, A.; Artim, L.; Charlton, S.; Dana, G.; Glenn, K.; Hunst, P.; Jennings, J.; Shilito, R.; Song, P. Sensitive PCR analysis of animal tissue samples for fragments of endogenous and transgenic plant DNA. J. Agric. Food Chem. 2004, 52, 6129–6135. [Google Scholar]
- Bottero, M.T.; Civera, T.; Nucera, D.; Rosati, S.; Sacchi, P.; Turi, R.M. A multiplex polymerase chain reaction for the identification of cows’, goat’s and sheep’s milk in dairy products. Int. Dairy J. 2003, 13, 277–282. [Google Scholar]
- Giannino, M.L.; Aliprandi, M.; Feligini, M.; Vanoni, L.; Brasca, M.; Fracchetti, F. A DNA array based assay for the characterization of microbial community in raw milk. J. Microbiol. Meth. 2009, 78, 181–188. [Google Scholar]
- Ponzoni, E.; Gianì, S.; Mastromauro, F.; Breviario, D. From milk to diet: feed recognition for milk authenticity. J. Dairy Sci. 2009, 92, 5583–5594. [Google Scholar]
- Bradley, A.; Green, M. Use and interpretation of somatic cell count data in dairy cows. In Practice 2005, 27, 310–315. [Google Scholar]
- Poms, R.E.; Hochsteiner, W.; Luger, K.; Glössl, J.; Foissy, H. Model studies on the detectability of genetically modified feeds in milk. J. Food Prot. 2003, 66, 304–310. [Google Scholar]
- Walker, G.P.; Dunshea, F.R.; Doyle, P.T. Effects of nutrition and management on the production and composition of milk fat and protein: a review. Aust. J. Agric. Res. 2004, 55, 1009–1028. [Google Scholar]
- Butler, G.; Stergiadis, S.; Eyre, M.; Leifert, C.; Borsari, A.; Canever, A.; Slots, T.; Nielsen, H.J. Effect of production system and geographic location on milk quality parameters. Proceedings of the 3rd QLIF Congress: Improving Sustainability in Organic and Low Input Food Production Systems, University of Hohenheim, Hohenheim, Germany, March 20-23, 2007; Available online: http://orgprints.org/10625/ (accessed in September 2009).
- Klotz, A.; Einspanier, R. Development of a DNA-based screening method to detect cow milk in ewe, goat and buffalo milk and dairy products using PCR-LCR-EIA-technique. Milchwissenschaft 2001, 56, 67–70. [Google Scholar]
© 2009 by the authors; licensee Molecular Diversity Preservation International, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Ponzoni, E.; Mastromauro, F.; Gianì, S.; Breviario, D. Traceability of Plant Diet Contents in Raw Cow Milk Samples. Nutrients 2009, 1, 251-262. https://doi.org/10.3390/nu1020251
Ponzoni E, Mastromauro F, Gianì S, Breviario D. Traceability of Plant Diet Contents in Raw Cow Milk Samples. Nutrients. 2009; 1(2):251-262. https://doi.org/10.3390/nu1020251
Chicago/Turabian StylePonzoni, Elena, Francesco Mastromauro, Silvia Gianì, and Diego Breviario. 2009. "Traceability of Plant Diet Contents in Raw Cow Milk Samples" Nutrients 1, no. 2: 251-262. https://doi.org/10.3390/nu1020251
APA StylePonzoni, E., Mastromauro, F., Gianì, S., & Breviario, D. (2009). Traceability of Plant Diet Contents in Raw Cow Milk Samples. Nutrients, 1(2), 251-262. https://doi.org/10.3390/nu1020251