Critical Evaluation of Gene Expression Changes in Human Tissues in Response to Supplementation with Dietary Bioactive Compounds: Moving Towards Better-Quality Studies
Abstract
:1. Introduction
2. Literature Searching, Studies Selection and Data Extraction
3. Summary of the Experimental Designs of the Intervention Trials Examined in this Review
4. Overview of Some Critical Issues Related to the RT-qPCR Experimental Protocols Used in the Intervention Trials
4.1. Sample Characterization and Handling
4.2. RNA Extraction Protocols, Quantity and Quality
4.3. Reference Genes
4.4. Gene Expression Data Reporting
4.5. Gene Expression Results: Association with Bioavailability of Bioactive Compounds and/or with Protein Confirmatory Studies
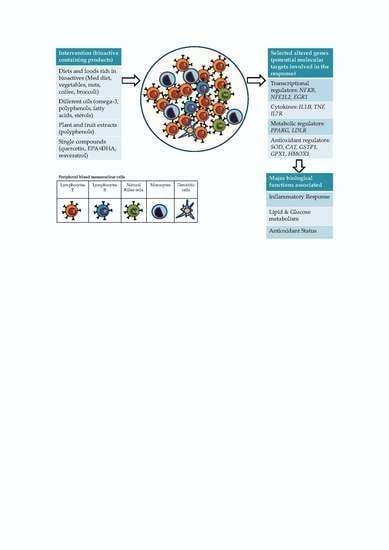
5. Gene Expression Changes in Human Cells and Tissues in Response to Food Bioactive Compounds: Overview of the Accumulated Evidence
5.1. Gene Expression Changes Specifically Reported in Blood Isolated Immune Cells in Response to Different Sources of Bioactive Compounds
5.2. Accumulated Evidence for Specific Gene Targets in Response to Different Sources of Bioactive Compounds
5.3. Summary of the Effects on Gene Expression of Specific Food Products Containing Bioactive Compounds
6. General Discussion
7. Concluding Remarks
Supplementary Materials
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Jayarathne, S.; Koboziev, I.; Park, O.H.; Oldewage-Theron, W.; Shen, C.L.; Moustaid-Moussa, N. Anti-inflammatory and anti-obesity properties of food bioactive components: Effects on adipose tissue. Prev. Nutr. Food Sci. 2017, 22, 251–262. [Google Scholar] [CrossRef] [PubMed]
- Giampieri, F.; Forbes-Hernandez, T.Y.; Gasparrini, M.; Afrin, S.; Cianciosi, D.; Reboredo-Rodriguez, P.; Varela-Lopez, A.; Quiles, J.L.; Mezzetti, B.; Battino, M. The health effects of strawberry bioactive compounds on molecular pathways related to chronic diseases. Ann. N. Y. Acad. Sci. 2017, 1398, 62–71. [Google Scholar] [CrossRef] [PubMed]
- García-Conesa, M.T. Dietary polyphenols against metabolic disorders: How far have we progressed in the understanding of the molecular mechanisms of action of these compounds? Crit. Rev. Food Sci. Nutr. 2017, 57, 1769–1786. [Google Scholar] [CrossRef] [PubMed]
- Nuñez-Sánchez, M.A.; González-Sarrías, A.; García-Villalba, R.; Monedero-Saiz, T.; García-Talavera, N.V.; Gómez-Sánchez, M.B.; Sánchez-Álvarez, C.; García-Albert, A.M.; Rodríguez-Gil, F.J.; Ruiz-Marín, M.; et al. Gene expression changes in colon tissues from colorectal cancer patients following the intake of an ellagitannin-containing pomegranate extract: A randomized clinical trial. J. Nutr. Biochem. 2017, 42, 126–133. [Google Scholar] [CrossRef] [PubMed]
- Tili, E.; Michaille, J.J. Promiscuous effects of some phenolic natural products on inflammation at least in part arise from their ability to modulate the expression of global regulators, namely microRNAs. Molecules 2016, 21, 1263. [Google Scholar] [CrossRef] [PubMed]
- Ali, F.; Ismail, A.; Kersten, S. Molecular mechanisms underlying the potential antiobesity-related diseases effect of cocoa polyphenols. Mol. Nutr. Food Res. 2014, 58, 33–48. [Google Scholar] [CrossRef] [PubMed]
- Gormaz, J.G.; Valls, N.; Sotomayor, C.; Turner, T.; Rodrigo, R. Potential role of polyphenols in the prevention of cardiovascular diseases: Molecular bases. Curr. Med. Chem. 2016, 23, 115–128. [Google Scholar] [CrossRef] [PubMed]
- Szwajgier, D.; Borowiec, K.; Pustelniak, K. The neuroprotective effects of phenolic acids: Molecular mechanism of action. Nutrients 2017, 9, 477. [Google Scholar] [CrossRef] [PubMed]
- Herrera-Marcos, L.V.; Lou-Bonafonte, J.M.; Arnal, C.; Navarro, M.A.; Osada, J. Transcriptomics and the Mediterranean diet: A systematic review. Nutrients 2017, 9, 472. [Google Scholar] [CrossRef] [PubMed]
- Rescigno, T.; Micolucci, L.; Tecce, M.F.; Capasso, A. Bioactive nutrients and nutrigenomics in age-related diseases. Molecules 2017, 22, 105. [Google Scholar] [CrossRef] [PubMed]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; Vandesompele, J.; Wittwer, C.T. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef] [PubMed]
- Bustin, S.; Nolan, T. Talking the talk, but not walking the walk: RT-qPCR as a paradigm for the lack of reproducibility in molecular research. Eur. J. Clin. Invest. 2017, 47, 756–774. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Derveaux, S.; Vandesompele, J.; Hellemans, J. How to do successful gene expression analysis using real-time PCR. Methods 2010, 50, 227–230. [Google Scholar] [CrossRef] [PubMed]
- Persson, I.; He, L.; Fang, C.; Normén, L.; Rylander, R. Influence of vegetables on the expression of GSTP1 in humans-a pilot intervention study (Sweden). Cancer Causes Control 2000, 11, 359–361. [Google Scholar] [CrossRef] [PubMed]
- Møller, P.; Vogel, U.; Pedersen, A.; Dragsted, L.O.; Sandström, B.; Loft, S. No effect of 600 grams fruit and vegetables per day on oxidative DNA damage and repair in healthy nonsmokers. Cancer Epidemiol. Biomarkers Prev. 2003, 12, 1016–1022. [Google Scholar] [PubMed]
- Almendingen, K.; Brevik, A.; Nymoen, D.A.; Hilmarsen, H.T.; Andresen, P.A.; Andersen, L.F.; Vatn, M. Modulation of COX-2 expression in peripheral blood cells by increased intake of fruit and vegetables? Eur. J. Clin. Nutr. 2005, 59, 597–602. [Google Scholar] [CrossRef] [PubMed]
- Dragsted, L.O.; Krath, B.; Ravn-Haren, G.; Vogel, U.B.; Vinggaard, A.M.; Bo Jensen, P.; Loft, S.; Rasmussen, S.E.; Sandstrom, T.; Pedersen, A. Biological effects of fruit and vegetables. Proc. Nutr. Soc. 2006, 65, 61–67. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Di Renzo, L.; Carraro, A.; Valente, R.; Iacopino, L.; Colica, C.; De Lorenzo, A. Intake of red wine in different meals modulates oxidized LDL level, oxidative and inflammatory gene expression in healthy people: A randomized crossover trial. Oxid. Med. Cell. Longev. 2014, 2014, 681318. [Google Scholar] [CrossRef] [PubMed]
- De Lorenzo, A.; Bernardini, S.; Gualtieri, P.; Cabibbo, A.; Perrone, M.A.; Giambini, I.; Di Renzo, L. Mediterranean meal versus Western meal effects on postprandial ox-LDL, oxidative and inflammatory gene expression in healthy subjects: A randomized controlled trial for nutrigenomic approach in cardiometabolic risk. Acta Diabetol. 2017, 54, 141–149. [Google Scholar] [CrossRef] [PubMed]
- Marques-Rocha, J.L.; Milagro, F.I.; Mansego, M.L.; Zulet, M.A.; Bressan, J.; Martínez, J.A. Expression of inflammation-related miRNAs in white blood cells from subjects with metabolic syndrome after 8 wk of following a Mediterranean diet-based weight loss program. Nutrition 2016, 32, 48–55. [Google Scholar] [CrossRef] [PubMed]
- Gasper, A.V.; Traka, M.; Bacon, J.R.; Smith, J.A.; Taylor, M.A.; Hawkey, C.J.; Barrett, D.A.; Mithen, R.F. Consuming broccoli does not induce genes associated with xenobiotic metabolism and cell cycle control in human gastric mucosa. J. Nutr. 2007, 137, 1718–1724. [Google Scholar] [CrossRef] [PubMed]
- Riedl, M.A.; Saxon, A.; Diaz-Sanchez, D. Oral sulforaphane increases Phase II antioxidant enzymes in the human upper airway. Clin. Immunol. 2009, 130, 244–251. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yanaka, A.; Fahey, J.W.; Fukumoto, A.; Nakayama, M.; Inoue, S.; Zhang, S.; Tauchi, M.; Suzuki, H.; Hyodo, I.; Yamamoto, M. Dietary sulforaphane-rich broccoli sprouts reduce colonization and attenuate gastritis in Helicobacter pylori-infected mice and humans. Cancer Prev. Res. 2009, 2, 353–360. [Google Scholar] [CrossRef] [PubMed]
- Riso, P.; Martini, D.; Møller, P.; Loft, S.; Bonacina, G.; Moro, M.; Porrini, M. DNA damage and repair activity after broccoli intake in young healthy smokers. Mutagenesis 2010, 25, 595–602. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Atwell, L.L.; Hsu, A.; Wong, C.P.; Stevens, J.F.; Bella, D.; Yu, T.W.; Pereira, C.B.; Löhr, C.V.; Christensen, J.M.; Dashwood, R.H.; et al. Absorption and chemopreventive targets of sulforaphane in humans following consumption of broccoli sprouts or a myrosinase-treated broccoli sprout extract. Mol. Nutr. Food Res. 2015, 59, 424–433. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Doss, J.F.; Jonassaint, J.C.; Garrett, M.E.; Ashley-Koch, A.E.; Telen, M.J.; Chi, J.T. Phase 1 study of a sulforaphane-containing broccoli sprout homogenate for sickle cell disease. PLoS ONE 2016, 11, e0152895. [Google Scholar] [CrossRef] [PubMed]
- Camargo, A.; Ruano, J.; Fernandez, J.M.; Parnell, L.D.; Jimenez, A.; Santos-Gonzalez, M.; Marin, C.; Perez-Martinez, P.; Uceda, M.; Lopez-Miranda, J.; et al. Gene expression changes in mononuclear cells in patients with metabolic syndrome after acute intake of phenol-rich virgin olive oil. BMC Genom. 2010, 11, 253. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Konstantinidou, V.; Covas, M.I.; Muñoz-Aguayo, D.; Khymenets, O.; de la Torre, R.; Saez, G.; Tormos Mdel, C.; Toledo, E.; Marti, A.; Ruiz-Gutiérrez, V.; et al. In vivo nutrigenomic effects of virgin olive oil polyphenols within the frame of the Mediterranean diet: A randomized controlled trial. FASEB J. 2010, 24, 2546–2557. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chan, J.M.; Weinberg, V.; Magbanua, M.J.; Sosa, E.; Simko, J.; Shinohara, K.; Federman, S.; Mattie, M.; Hughes-Fulford, M.; Haqq, C.; et al. Nutritional supplements, COX-2 and IGF-1 expression in men on active surveillance for prostate cancer. Cancer Causes Control 2011, 22, 141–150. [Google Scholar] [CrossRef] [PubMed]
- Castañer, O.; Covas, M.I.; Khymenets, O.; Nyyssonen, K.; Konstantinidou, V.; Zunft, H.F.; de la Torre, R.; Muñoz-Aguayo, D.; Vila, J.; Fitó, M. Protection of LDL from oxidation by olive oil polyphenols is associated with a downregulation of CD40-ligand expression and its downstream products in vivo in humans. Am. J. Clin. Nutr. 2012, 95, 1238–1244. [Google Scholar] [Green Version]
- Farràs, M.; Valls, R.M.; Fernández-Castillejo, S.; Giralt, M.; Solà, R.; Subirana, I.; Motilva, M.J.; Konstantinidou, V.; Covas, M.I.; Fitó, M. Olive oil polyphenols enhance the expression of cholesterol efflux related genes in vivo in humans. A randomized controlled trial. J. Nutr. Biochem. 2013, 24, 1334–1339. [Google Scholar] [CrossRef] [PubMed]
- Perez-Herrera, A.; Rangel-Zuñiga, O.A.; Delgado-Lista, J.; Marin, C.; Perez-Martinez, P.; Tasset, I.; Tunez, I.; Quintana-Navarro, G.M.; Lopez-Segura, F.; Luque de Castro, M.D.; et al. The antioxidants in oils heated at frying temperature, whether natural or added, could protect against postprandial oxidative stress in obese people. Food Chem. 2013, 138, 2250–2259. [Google Scholar] [CrossRef] [PubMed]
- Rangel-Zuñiga, O.A.; Haro, C.; Perez-Martinez, P.; Delgado-Lista, J.; Marin, C.; Quintana-Navarro, G.M.; Tinahones, F.J.; Malagón, M.M.; Lopez-Segura, F.; López-Miranda, J.; Perez-Jimenez, F.; Camargo, A. Effect of frying oils on the postprandial endoplasmic reticulum stress in obese people. Mol. Nutr. Food Res. 2014, 58, 2239–2242. [Google Scholar] [CrossRef] [PubMed]
- Hernáez, Á.; Remaley, A.T.; Farràs, M.; Fernández-Castillejo, S.; Subirana, I.; Schröder, H.; Fernández-Mampel, M.; Muñoz-Aguayo, D.; Sampson, M.; Solà, R.; et al. Olive oil polyphenols decrease LDL concentrations and LDL atherogenicity in men in a randomized controlled trial. J. Nutr. 2015, 145, 1692–1697. [Google Scholar] [CrossRef] [PubMed]
- Martín-Peláez, S.; Castañer, O.; Konstantinidou, V.; Subirana, I.; Muñoz-Aguayo, D.; Blanchart, G.; Gaixas, S.; de la Torre, R.; Farré, M.; Sáez, G.T.; et al. Effect of olive oil phenolic compounds on the expression of blood pressure-related genes in healthy individuals. Eur. J. Nutr. 2017, 56, 663–670. [Google Scholar] [CrossRef] [PubMed]
- Kruse, M.; von Loeffelholz, C.; Hoffmann, D.; Pohlmann, A.; Seltmann, A.C.; Osterhoff, M.; Hornemann, S.; Pivovarova, O.; Rohn, S.; Jahreis, G.; et al. Dietary rapeseed/canola-oil supplementation reduces serum lipids and liver enzymes and alters postprandial inflammatory responses in adipose tissue compared to olive-oil supplementation in obese men. Mol. Nutr. Food Res. 2015, 59, 507–519. [Google Scholar] [CrossRef] [PubMed]
- González-Sarrías, A.; Giménez-Bastida, J.A.; García-Conesa, M.T.; Gómez-Sánchez, M.B.; García-Talavera, N.V.; Gil-Izquierdo, A.; Sánchez-Alvarez, C.; Fontana-Compiano, L.O.; Morga-Egea, J.P.; Pastor-Quirante, F.A.; et al. Occurrence of urolithins, gut microbiota ellagic acid metabolites and proliferation markers expression response in the human prostate gland upon consumption of walnuts and pomegranate juice. Mol. Nutr. Food Res. 2010, 54, 311–322. [Google Scholar] [CrossRef] [PubMed]
- Hernández-Alonso, P.; Salas-Salvadó, J.; Baldrich-Mora, M.; Juanola-Falgarona, M.; Bulló, M. Beneficial effect of pistachio consumption on glucose metabolism, insulin resistance, inflammation, and related metabolic risk markers: A randomized clinical trial. Diabetes Care 2014, 37, 3098–3105. [Google Scholar] [CrossRef] [PubMed]
- Donadio, J.L.S.; Rogero, M.M.; Cockell, S.; Hesketh, J.; Cozzolino, S.M.F. Influence of genetic variations in selenoprotein genes on the pattern of gene expression after supplementation with Brazil nuts. Nutrients 2017, 9, 739. [Google Scholar] [CrossRef] [PubMed]
- Di Renzo, L.; Merra, G.; Botta, R.; Gualtieri, P.; Manzo, A.; Perrone, M.A.; Mazza, M.; Cascapera, S.; De Lorenzo, A. Post-prandial effects of hazelnut-enriched high fat meal on LDL oxidative status, oxidative and inflammatory gene expression of healthy subjects: A randomized trial. Eur. Rev. Med. Pharmacol. Sci. 2017, 21, 1610–1626. [Google Scholar] [PubMed]
- Guarrera, S.; Sacerdote, C.; Fiorini, L.; Marsala, R.; Polidoro, S.; Gamberini, S.; Saletta, F.; Malaveille, C.; Talaska, G.; Vineis, P.; et al. Expression of DNA repair and metabolic genes in response to a flavonoid-rich diet. Br. J. Nutr. 2007, 98, 525–533. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Volz, N.; Boettler, U.; Winkler, S.; Teller, N.; Schwarz, C.; Bakuradze, T.; Eisenbrand, G.; Haupt, L.; Griffiths, L.R.; Stiebitz, H.; et al. Effect of coffee combining green coffee bean constituents with typical roasting products on the Nrf2/ARE pathway in vitro and in vivo. J. Agric. Food Chem. 2012, 60, 9631–9641. [Google Scholar] [CrossRef] [PubMed]
- Boettler, U.; Volz, N.; Teller, N.; Haupt, L.M.; Bakuradze, T.; Eisenbrand, G.; Bytof, G.; Lantz, I.; Griffiths, L.R.; Marko, D. Induction of antioxidative Nrf2 gene transcription by coffee in humans: depending on genotype? Mol. Biol. Rep. 2012, 39, 7155–7162. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Pascual-Teresa, S.; Johnston, K.L.; DuPont, M.S.; O’Leary, K.A.; Needs, P.W.; Morgan, L.M.; Clifford, M.N.; Bao, Y.; Williamson, G. Quercetin metabolites downregulate cyclooxygenase-2 transcription in human lymphocytes ex vivo but not in vivo. J. Nutr. 2004, 134, 552–557. [Google Scholar] [CrossRef] [PubMed]
- Marotta, F.; Koike, K.; Lorenzetti, A.; Jain, S.; Signorelli, P.; Metugriachuk, Y.; Mantello, P.; Locorotondo, N. Regulating redox balance gene expression in healthy individuals by nutraceuticals: A pilot study. Rejuvenation Res. 2010, 13, 175–178. [Google Scholar] [CrossRef] [PubMed]
- Bertuccelli, G.; Zerbinati, N.; Marcellino, M.; Nanda Kumar, N.S.; He, F.; Tsepakolenko, V.; Cervi, J.; Lorenzetti, A.; Marotta, F. Effect of a quality-controlled fermented nutraceutical on skin aging markers: An antioxidant-control, double-blind study. Exp. Ther. Med. 2016, 11, 909–916. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ishikawa, H.; Goto, M.; Matsuura, N.; Murakami, Y.; Goto, C.; Sakai, T.; Kanazawa, K. A pilot, randomized, placebo-controlled, double-blind phase 0/biomarker study on effect of artepillin C-rich extract of Brazilian propolis in frequent colorectal adenoma polyp patients. J. Am. Coll. Nutr. 2012, 31, 327–337. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, A.V.; Martinez, M.; Stamos, M.J.; Moyer, M.P.; Planutis, K.; Hope, C.; Holcombe, R.F. Results of a phase I pilot clinical trial examining the effect of plant-derived resveratrol and grape powder on Wnt pathway target gene expression in colonic mucosa and colon cancer. Cancer Manag. Res. 2009, 1, 25–37. [Google Scholar] [PubMed]
- Weseler, A.R.; Ruijters, E.J.; Drittij-Reijnders, M.J.; Reesink, K.D.; Haenen, G.R.; Bast, A. Pleiotropic benefit of monomeric and oligomeric flavanols on vascular health—A randomized controlled clinical pilot study. PLoS ONE 2011, 6, e28460. [Google Scholar] [CrossRef] [PubMed]
- Barona, J.; Blesso, C.N.; Andersen, C.J.; Park, Y.; Lee, J.; Fernandez, M.L. Grape consumption increases anti-inflammatory markers and upregulates peripheral nitric oxide synthase in the absence of dyslipidemias in men with metabolic syndrome. Nutrients 2012, 4, 1945–1957. [Google Scholar] [CrossRef] [PubMed]
- Tomé-Carneiro, J.; Larrosa, M.; Yáñez-Gascón, M.J.; Dávalos, A.; Gil-Zamorano, J.; Gonzálvez, M.; García-Almagro, F.J.; Ruiz Ros, J.A.; Tomás-Barberán, F.A.; Espín, J.C.; et al. One-year supplementation with a grape extract containing resveratrol modulates inflammatory-related microRNAs and cytokines expression in peripheral blood mononuclear cells of type 2 diabetes and hypertensive patients with coronary artery disease. Pharmacol. Res. 2013, 72, 69–82. [Google Scholar] [CrossRef] [PubMed]
- Mallery, S.R.; Zwick, J.C.; Pei, P.; Tong, M.; Larsen, P.E.; Shumway, B.S.; Lu, B.; Fields, H.W.; Mumper, R.J.; Stoner, G.D. Topical application of a bioadhesive black raspberry gel modulates gene expression and reduces cyclooxygenase 2 protein in human premalignant oral lesions. Cancer Res. 2008, 68, 4945–4957. [Google Scholar] [CrossRef] [PubMed]
- Kropat, C.; Mueller, D.; Boettler, U.; Zimmermann, K.; Heiss, E.H.; Dirsch, V.M.; Rogoll, D.; Melcher, R.; Richling, E.; Marko, D. Modulation of Nrf2-dependent gene transcription by bilberry anthocyanins in vivo. Mol. Nutr. Food Res. 2013, 57, 545–550. [Google Scholar] [CrossRef] [PubMed]
- Knobloch, T.J.; Uhrig, L.K.; Pearl, D.K.; Casto, B.C.; Warner, B.M.; Clinton, S.K.; Sardo-Molmenti, C.L.; Ferguson, J.M.; Daly, B.T.; Riedl, K.; et al. Suppression of proinflammatory and prosurvival biomarkers in oral cancer patients consuming ablack raspberry phytochemical-rich troche. Cancer Prev. Res. 2016, 9, 159–171. [Google Scholar] [CrossRef] [PubMed]
- Xie, L.; Vance, T.; Kim, B.; Lee, S.G.; Caceres, C.; Wang, Y.; Hubert, P.A.; Lee, J.Y.; Chun, O.K.; Bolling, B.W. Aronia berry polyphenol consumption reduces plasma total and low-density lipoprotein cholesterol in former smokers without lowering biomarkers of inflammation and oxidative stress: a randomized controlled trial. Nutr. Res. 2017, 37, 67–77. [Google Scholar] [CrossRef] [PubMed]
- Knott, A.; Reuschlein, K.; Mielke, H.; Wensorra, U.; Mummert, C.; Koop, U.; Kausch, M.; Kolbe, L.; Peters, N.; Stäb, F.; et al. Natural Arctium lappa fruit extract improves the clinical signs of aging skin. J. Cosmet. Dermatol. 2008, 7, 281–289. [Google Scholar] [CrossRef] [PubMed]
- Shrestha, S.; Freake, H.C.; McGrane, M.M.; Volek, J.S.; Fernandez, M.L. A combination of psyllium and plant sterols alters lipoprotein metabolism in hypercholesterolemic subjects by modifying the intravascular processing of lipoproteins and increasing LDL uptake. J. Nutr. 2007, 137, 1165–1170. [Google Scholar] [CrossRef] [PubMed]
- Ghanim, H.; Sia, C.L.; Abuaysheh, S.; Korzeniewski, K.; Patnaik, P.; Marumganti, A.; Chaudhuri, A.; Dandona, P. An antiinflammatory and reactive oxygen species suppressive effects of an extract of Polygonum cuspidatum containing resveratrol. J. Clin. Endocrinol. Metab. 2010, 95, E1–E8. [Google Scholar] [CrossRef] [PubMed]
- Marini, A.; Grether-Beck, S.; Jaenicke, T.; Weber, M.; Burki, C.; Formann, P.; Brenden, H.; Schönlau, F.; Krutmann, J. Pycnogenol® effects on skin elasticity and hydration coincide with increased gene expressions of collagen type I and hyaluronic acid synthase in women. Skin Pharmacol. Physiol. 2012, 25, 86–92. [Google Scholar] [CrossRef] [PubMed]
- Carrera-Quintanar, L.; Funes, L.; Vicente-Salar, N.; Blasco-Lafarga, C.; Pons, A.; Micol, V.; Roche, E. Effect of polyphenol supplements on redox status of blood cells: A randomized controlled exercise training trial. Eur. J. Nutr. 2015, 54, 1081–1093. [Google Scholar] [CrossRef] [PubMed]
- Turowski, J.B.; Pietrofesa, R.A.; Lawson, J.A.; Christofidou-Solomidou, M.; Hadjiliadis, D. Flaxseed modulates inflammatory and oxidative stress biomarkers in cystic fibrosis: A pilot study. BMC Complement. Altern. Med. 2015, 15, 148. [Google Scholar] [CrossRef] [PubMed]
- Plat, J.; Mensink, R.P. Effects of plant stanol esters on LDL receptor protein expression and on LDL receptor and HMG-CoA reductase mRNA expression in mononuclear blood cells of healthy men and women. FASEB J. 2002, 16, 258–260. [Google Scholar] [CrossRef] [PubMed]
- Crespo, M.C.; Tomé-Carneiro, J.; Burgos-Ramos, E.; Loria Kohen, V.; Espinosa, M.I.; Herranz, J.; Visioli, F. One-week administration of hydroxytyrosol to humans does not activate Phase II enzymes. Pharmacol. Res. 2015, 95–96, 132–137. [Google Scholar] [CrossRef] [PubMed]
- Boss, A.; Kao, C.H.; Murray, P.M.; Marlow, G.; Barnett, M.P.; Ferguson, L.R. Human intervention study to assess the effects of supplementation with olive leaf extract on peripheral blood mononuclear cell gene expression. Int. J. Mol Sci. 2016, 17, 2019. [Google Scholar] [CrossRef] [PubMed]
- Daak, A.A.; Elderdery, A.Y.; Elbashir, L.M.; Mariniello, K.; Mills, J.; Scarlett, G.; Elbashir, M.I.; Ghebremeskel, K. Omega 3 (n-3) fatty acids down-regulate nuclear factor-kappa B (NF-KB) gene and blood cell adhesion molecule expression in patients with homozygous sickle cell disease. Blood Cells Mol. Dis. 2015, 55, 48–55. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Labonté, M.È.; Couture, P.; Tremblay, A.J.; Hogue, J.C.; Lemelin, V.; Lamarche, B. Eicosapentaenoic and docosahexaenoic acid supplementation and inflammatory gene expression in the duodenum of obese patients with type 2 diabetes. Nutr. J. 2013, 12, 98. [Google Scholar] [CrossRef] [PubMed]
- Jamilian, M.; Samimi, M.; Mirhosseini, N.; Afshar Ebrahimi, F.; Aghadavod, E.; Taghizadeh, M.; Asemi, Z. A randomized double-blinded, placebo-controlled trial investigating the effect of fish oil supplementation on gene expression related to insulin action, blood lipids, and inflammation in gestational diabetes mellitus-fish oil supplementation and gestational diabetes. Nutrients 2018, 10, 163. [Google Scholar] [CrossRef] [PubMed]
- Marini, A.; Jaenicke, T.; Grether-Beck, S.; Le Floc’h, C.; Cheniti, A.; Piccardi, N.; Krutmann, J. Prevention of polymorphic light eruption by oral administration of a nutritional supplement containing lycopene, β-carotene, and Lactobacillus johnsonii: Results from a randomized, placebo-controlled, double-blinded study. Photodermatol. Photoimmunol. Photomed. 2014, 30, 189–194. [Google Scholar] [CrossRef] [PubMed]
- Farris, P.; Yatskayer, M.; Chen, N.; Krol, Y.; Oresajo, C. Evaluation of efficacy and tolerance of a nighttime topical antioxidant containing resveratrol, baicalin, and vitamin E for treatment of mild to moderately photodamaged skin. J. Drugs Dermatol. 2014, 13, 1467–1472. [Google Scholar] [PubMed]
- Radler, U.; Stangl, H.; Lechner, S.; Lienbacher, G.; Krepp, R.; Zeller, E.; Brachinger, M.; Eller-Berndl, D.; Fischer, A.; Anzur, C.; et al. A combination of (ω-3) polyunsaturated fatty acids, polyphenols and l-carnitine reduces the plasma lipid levels and increases the expression of genes involved in fatty acid oxidation in human peripheral blood mononuclear cells and HepG2 cells. Ann. Nutr. Metab. 2011, 58, 133–140. [Google Scholar] [CrossRef] [PubMed]
- Frommel, T.O.; Lietz, H.; Mobarhan, S. Expression of mRNA for the gap-junctional protein connexin43 in human colonic tissue is variable in response to beta-carotene supplementation. Nutr. Cancer 1994, 22, 257–265. [Google Scholar] [CrossRef] [PubMed]
- Vors, C.; Allaire, J.; Marin, J.; Lépine, M.C.; Charest, A.; Tchernof, A.; Couture, P.; Lamarche, B. Inflammatory gene expression in whole blood cells after EPA vs. DHA supplementation: Results from the ComparED study. Atherosclerosis 2017, 257, 116–122. [Google Scholar] [CrossRef] [PubMed]
- Yoshino, J.; Conte, C.; Fontana, L.; Mittendorfer, B.; Imai, S.; Schechtman, K.B.; Gu, C.; Kunz, I.; Rossi Fanelli, F.; Patterson, B.W.; Klein, S. Resveratrol supplementation does not improve metabolic function in nonobese women with normal glucose tolerance. Cell Metab. 2012, 16, 658–664. [Google Scholar] [CrossRef] [PubMed]
- Poulsen, M.M.; Vestergaard, P.F.; Clasen, B.F.; Radko, Y.; Christensen, L.P.; Stødkilde-Jørgensen, H.; Møller, N.; Jessen, N.; Pedersen, S.B.; Jørgensen, J.O. High-dose resveratrol supplementation in obese men: An investigator-initiated, randomized, placebo-controlled clinical trial of substrate metabolism, insulin sensitivity, and body composition. Diabetes 2013, 62, 1186–1195. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Olesen, J.; Gliemann, L.; Biensø, R.; Schmidt, J.; Hellsten, Y.; Pilegaard, H. Exercise training, but not resveratrol, improves metabolic and inflammatory status in skeletal muscle of aged men. J. Physiol. 2014, 592, 1873–1886. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chachay, V.S.; Macdonald, G.A.; Martin, J.H.; Whitehead, J.P.; O’Moore-Sullivan, T.M.; Lee, P.; Franklin, M.; Klein, K.; Taylor, P.J.; Ferguson, M.; et al. Resveratrol does not benefit patients with nonalcoholic fatty liver disease. Clin. Gastroenterol. Hepatol. 2014, 12, 2092–2103. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yiu, E.M.; Tai, G.; Peverill, R.E.; Lee, K.J.; Croft, K.D.; Mori, T.A.; Scheiber-Mojdehkar, B.; Sturm, B.; Praschberger, M.; Vogel, A.P.; et al. An open-label trial in Friedreich ataxia suggests clinical benefit with high-dose resveratrol, without effect on frataxin levels. J. Neurol. 2015, 262, 1344–1353. [Google Scholar] [CrossRef] [PubMed]
- Mansur, A.P.; Roggerio, A.; Goes, M.F.S.; Avakian, S.D.; Leal, D.P.; Maranhão, R.C.; Strunz, C.M.C. Serum concentrations and gene expression of sirtuin 1 in healthy and slightly overweight subjects after caloric restriction or resveratrol supplementation: A randomized trial. Int. J. Cardiol. 2017, 227, 788–794. [Google Scholar] [CrossRef] [PubMed]
- Morrow, D.M.; Fitzsimmons, P.E.; Chopra, M.; McGlynn, H. Dietary supplementation with the anti-tumour promoter quercetin: Its effects on matrix metalloproteinase gene regulation. Mutat. Res. 2001, 480–481, 269–276. [Google Scholar] [CrossRef]
- Nieman, D.C.; Henson, D.A.; Davis, J.M.; Angela Murphy, E.; Jenkins, D.P.; Gross, S.J.; Carmichael, M.D.; Quindry, J.C.; Dumke, C.L.; Utter, A.C.; et al. Quercetin’s influence on exercise-induced changes in plasma cytokines and muscle and leukocyte cytokine mRNA. J. Appl. Physiol. 2007, 103, 1728–1735. [Google Scholar] [CrossRef] [PubMed]
- Boesch-Saadatmandi, C.; Pospissil, R.T.; Graeser, A.C.; Canali, R.; Boomgaarden, I.; Doering, F.; Wolffram, S.; Egert, S.; Mueller, M.J.; Rimbach, G. Effect of quercetin on paraoxonase 2 levels in RAW264.7 macrophages and in human monocytes—Role of quercetin metabolism. Int. J. Mol. Sci. 2009, 10, 4168–4177. [Google Scholar] [CrossRef] [PubMed]
- Nelson, D.M.; Joseph, B.; Hillion, J.; Segal, J.; Karp, J.E.; Resar, L.M. Flavopiridol induces BCL-2 expression and represses oncogenic transcription factors in leukemic blasts from adults with refractory acute myeloid leukemia. Leuk. Lymphoma 2011, 52, 1999–2006. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lazarevic, B.; Hammarström, C.; Yang, J.; Ramberg, H.; Diep, L.M.; Karlsen, S.J.; Kucuk, O.; Saatcioglu, F.; Taskèn, K.A.; Svindland, A. The effects of short-term genistein intervention on prostate biomarker expression in patients with localised prostate cancer before radical prostatectomy. Br. J. Nutr. 2012, 108, 2138–2147. [Google Scholar] [CrossRef] [PubMed]
- Kerksick, C.M.; Roberts, M.D.; Dalbo, V.J.; Kreider, R.B.; Willoughby, D.S. Changes in skeletal muscle proteolytic gene expression after prophylactic supplementation of EGCG and NAC and eccentric damage. Food Chem. Toxicol. 2013, 61, 47–52. [Google Scholar] [CrossRef] [PubMed]
- Most, J.; van Can, J.G.; van Dijk, J.W.; Goossens, G.H.; Jocken, J.; Hospers, J.J.; Bendik, I.; Blaak, E.E. A 3-day EGCG-supplementation reduces interstitial lactate concentration in skeletal muscle of overweight subjects. Sci. Rep. 2015, 5, 17896. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koosirirat, C.; Linpisarn, S.; Changsom, D.; Chawansuntati, K.; Wipasa, J. Investigation of the anti-inflammatory effect of Curcuma longa in Helicobacter pylori-infected patients. Int. Immunopharmacol. 2010, 10, 815–818. [Google Scholar] [CrossRef] [PubMed]
- Klickovic, U.; Doberer, D.; Gouya, G.; Aschauer, S.; Weisshaar, S.; Storka, A.; Bilban, M.; Wolzt, M. Human pharmacokinetics of high dose oral curcumin and its effect on heme oxygenase-1 expression in healthy male subjects. Biomed Res. Int. 2014, 2014, 458592. [Google Scholar] [CrossRef] [PubMed]
- Stolberg, H.O.; Norman, G.; Trop, I. Randomized controlled trials. AJR Am. J. Roentgenol. 2004, 183, 1539–1544. [Google Scholar] [CrossRef] [PubMed]
- Stoney, C.M.; Johnson, L.L. Design of clinical studies and trials. In Principles and Practice of Clinical Research, 3rd ed.; Gallin, J.I., Ognibene, P.F., Eds.; Elsevier Inc.: San Diego, CA, USA, 2012; Chapter 19; pp. 225–243. ISBN 978-0-12-382167-6. [Google Scholar]
- Schroeder, A.; Mueller, O.; Stocker, S.; Salowsky, R.; Leiber, M.; Gassmann, M.; Lightfoot, S.; Menzel, W.; Granzow, M.; Ragg, T. The RIN: An RNA integrity number for assigning integrity values to RNA measurements. BMC Mol. Biol. 2006, 7, 3. [Google Scholar] [CrossRef] [PubMed]
- Chapman, J.R.; Waldenström, J. With reference to reference genes: A systematic review of endogenous controls in gene expression studies. PLoS ONE 2015, 10, e0141853. [Google Scholar] [CrossRef] [PubMed]
- GeneCards. Available online: http://www.genecards.org/.
- Akamine, R.; Yamamoto, T.; Watanabe, M.; Yamazaki, N.; Kataoka, M.; Ishikawa, M.; Ooie, T.; Baba, Y.; Shinohara, Y. Usefulness of the 5’ region of the cDNA encoding acidic ribosomal phosphoprotein P0 conserved among rats, mice, and humans as a standard probe for gene expression analysis in different tissues and animal species. J. Biochem. Biophys. Methods 2007, 70, 481–486. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- McMahon, S.B.; Monroe, J.G. The role of early growth response gene 1 (egr-1) in regulation of the immune response. J. Leukoc. Biol. 1996, 60, 159–166. [Google Scholar] [CrossRef] [PubMed]
- Chedea, V.S.; Vicaş, S.I.; Sticozzi, C.; Pessina, F.; Frosini, M.; Maioli, E.; Valacchi, G. Resveratrol: From diet to topical usage. Food Funct. 2017, 8, 3879–3892. [Google Scholar] [CrossRef] [PubMed]
- Treviño-Saldaña, N.; García-Rivas, G. Regulation of sirtuin-mediated protein deacetylation by cardioprotective phytochemicals. Oxid. Med. Cell. Longev. 2017, 2017, 1750306. [Google Scholar] [CrossRef] [PubMed]
- De Mello, V.D.; Kolehmanien, M.; Schwab, U.; Pulkkinen, L.; Uusitupa, M. Gene expression of peripheral blood mononuclear cells as a tool in dietary intervention studies: What do we know so far? Mol. Nutr. Food Res. 2012, 56, 1160–1172. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schmidt, M.V.; Brüne, B.; von Knethen, A. The nuclear hormone receptor PPARγ as a therapeutic target in major diseases. Sci. World J. 2010, 10, 2181–2197. [Google Scholar] [CrossRef] [PubMed]
- Manach, C.; Milenkovic, D.; Van de Wiele, T.; Rodriguez-Mateos, A.; de Roos, B.; Garcia-Conesa, M.T.; Landberg, R.; Gibney, E.R.; Heinonen, M.; Tomás-Barberán, F.; et al. Addressing the inter-individual variation in response to consumption of plant food bioactives: Towards a better understanding of their role in healthy aging and cardiometabolic risk reduction. Mol. Nutr. Food Res. 2017, 61. [Google Scholar] [CrossRef] [PubMed]
- Fleige, S.; Pfaffl, M.W. RNA integrity and the effect on the real-time qRT-PCR performance. Mol. Aspects Med. 2006, 27, 126–139. [Google Scholar] [CrossRef] [PubMed]
- Altman, D.G.; Bland, J.M. Standard deviations and standard errors. BMJ 2005, 331, 903. [Google Scholar] [CrossRef] [PubMed]
- Cumming, G. The new statistics: Why and how. Psychol. Sci. 2014, 25, 7–29. [Google Scholar] [CrossRef] [PubMed]
- Sadeghabadi, Z.A.; Nourbakhsh, M.; Alaee, M.; Larijani, B.; Razzaghy-Azar, M. Peroxisome proliferator-activated receptor gamma expression in peripheral blood mononuclear cells and angiopoietin-like protein 4 levels in obese children and adolescents. J. Endocrinol. Invest. 2018, 41, 241–247. [Google Scholar] [CrossRef] [PubMed]
- Oliver, P.; Reynés, B.; Caimari, A.; Palou, A. Peripheral blood mononuclear cells: A potential source of homeostatic imbalance markers associated with obesity development. Pflugers Arch. 2013, 465, 459–468. [Google Scholar] [CrossRef] [PubMed]
- Viguerie, N.; Vidal, H.; Arner, P.; Holst, C.; Verdich, C.; Avizou, S.; Astrup, A.; Saris, W.H.; Macdonald, I.A.; Klimcakova, E.; et al. Adipose tissue gene expression in obese subjects during low-fat and high-fat hypocaloric diets. Diabetologia 2005, 48, 123–131. [Google Scholar] [CrossRef] [PubMed]
- Gupta, S.C.; Tyagi, A.K.; Deshmukh-Taskar, P.; Hinojosa, M.; Prasad, S.; Aggarwal, B.B. Downregulation of tumor necrosis factor and other proinflammatory biomarkers by polyphenols. Arch. Biochem. Biophys. 2014, 559, 91–99. [Google Scholar] [CrossRef] [PubMed]
- Sahebkar, A.; Cicero, A.F.G.; Simental-Mendía, L.E.; Aggarwal, B.B.; Gupta, S.C. Curcumin downregulates human tumor necrosis factor-α levels: A systematic review and meta-analysis of randomized controlled trials. Pharmacol. Res. 2016, 107, 234–242. [Google Scholar] [CrossRef] [PubMed]
- Schwager, J.; Richard, N.; Widmer, F.; Raederstorff, D. Resveratrol distinctively modulates the inflammatory profiles of immune and endothelial cells. BMC Complement. Altern. Med. 2017, 17, 309. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Timmers, S.; Konings, E.; Bilet, L.; Houtkooper, R.H.; van de Weijer, T.; Goossens, G.H.; Hoeks, J.; van der Krieken, S.; Ryu, D.; Kersten, S.; et al. Calorie restriction-like effects of 30 days of resveratrol supplementation on energy metabolism and metabolic profile in obese humans. Cell Metab. 2011, 14, 612–622. [Google Scholar] [CrossRef] [PubMed]
- Hussain, T.; Tan, B.; Yin, Y.; Blachier, F.; Tossou, M.C.; Rahu, N. Oxidative stress and inflammation: What polyphenols can do for us? Oxid. Med. Cell. Longev. 2016, 2016, 7432797. [Google Scholar] [CrossRef] [PubMed]
- Lei, X.G.; Cheng, W.H.; McClung, J.P. Metabolic regulation and function of glutathione peroxidase-1. Annu. Rev. Nutr. 2007, 27, 41–61. [Google Scholar] [CrossRef] [PubMed]
- Gouvinhas, I.; Machado, N.; Sobreira, C.; Domínguez-Perles, R.; Gomes, S.; Rosa, E.; Barros, A.I.R.N.A. Critical review on the significance of olive phytochemicals in plant physiology and human health. Molecules 2017, 22, 1986. [Google Scholar] [CrossRef] [PubMed]
- Verkerk, R.; Schreiner, M.; Krumbein, A.; Ciska, E.; Holst, B.; Rowland, I.; De Schrijver, R.; Hansen, M.; Gerhäuser, C.; Mithen, R.; Dekker, M. Glucosinolates in Brassica vegetables: The influence of the food supply chain on intake, bioavailability and human health. Mol. Nutr. Food Res. 2009, 53, S219. [Google Scholar] [CrossRef] [PubMed]
- Pagel, J.I.; Deindl, E. Early growth response 1—A transcription factor in the crossfire of signal transduction cascades. Indian J. Biochem. Biophys. 2011, 48, 226–235. [Google Scholar] [PubMed]
- Della Ragione, F.; Cucciolla, V.; Criniti, V.; Indaco, S.; Borriello, A.; Zappia, V. Antioxidants induce different phenotypes by a distinct modulation of signal transduction. FEBS Lett. 2002, 532, 289–294. [Google Scholar] [CrossRef] [Green Version]
- Cornblatt, B.S.; Ye, L.; Dinkova-Kostova, A.T.; Erb, M.; Fahey, J.W.; Singh, N.K.; Chen, M.S.; Stierer, T.; Garrett-Mayer, E.; Argani, P.; et al. Preclinical and clinical evaluation of sulforaphane for chemoprevention in the breast. Carcinogenesis 2007, 28, 1485–1490. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Núñez-Sánchez, M.A.; González-Sarrías, A.; Romo-Vaquero, M.; García-Villalba, R.; Selma, M.V.; Tomás-Barberán, F.A.; García-Conesa, M.T.; Espín, J.C. Dietary phenolics against colorectal cancer—From promising preclinical results to poor translation into clinical trials: Pitfalls and future needs. Mol. Nutr. Food Res. 2015, 59, 1274–1291. [Google Scholar] [CrossRef] [PubMed]

| Gene Symbol | n Studies 1 | Cell/Tissue Samples in Which the Gene Has Been Used as a Reference Gene |
|---|---|---|
| Most common genes used as reference genes | ||
| GAPDH | 31 [4,16,18,20,25,26,28,32,33,34,35,37,39,40,44,45,46,49,50,51,53,55,57,64,65,67,72,73,78,79,86] 1 tested, but not used [63] | Blood, white blood cells, mononuclear cells, lymphocytes, neutrophils, gastric antrum, colon cancer and colon normal tissue, prostate hyperplasia and prostate cancer tissue, skeletal muscle, adipose tissues |
| ACTB | 16 [14,18,40,41,42,43,46,48,58,63,64,70,76,81,82,84] 1 tested, but not used [53] | Blood, white blood cells, leukemic blasts, lymphocytes, CD14+ monocytes, colon cancer and colon normal tissue, skeletal muscle tissue, skin tissue |
| 18S rRNA | 11 [15,17,21,22,24,56,59,61,80,85,87] 1 considered, but not used [66] | White blood cells, mononuclear cells, skeletal muscle tissue, buccal swabs, gastric mucosa, adipose tissue, nasal cells, epidermis blister, buttock skin |
| Other genes less commonly used as reference genes | ||
| B2M | 6 [18,40,70,71,74,82] | Blood, mononuclear cells, leukemic blasts, colon tissue, skeletal muscle, adipose tissue |
| HPRT1 | 3 [18,38,40], 2 tested, but not used [4,72] 1 considered, but not used [66] | Blood, lymphocytes |
| GUSB | 2 [29,52], 1 tested, but not used [4] | Oral mucosa, prostate tissue |
| RPLP0 | 3 [40,60,82], 1 tested, but not used [63] | Blood, leukemic blasts, lymphocytes, neutrophils |
| RPL13A | 1 [27] | Mononuclear cells |
| RPL32 | 1 [36] | Adipose tissue |
| HMBS | 2 [45,47] | Neutrophils, colon mucosa |
| ATP5O | 1 [66] | Duodenal biopsies |
| DUSP1 | 1 [54] | Oral biopsies |
| ALAS1 | 1 [83] | Prostate cancer and normal tissue |
| YWHAZ | 1 [38] | Lymphocytes |
| UBC | 1 [58] | Mononuclear cells |
| PPIA | 1 [58] | Mononuclear cells |
| G6PD | 2 [66,72] | Blood (tested but not used), duodenal tissue (selected but not used) |
| Other reference molecules used | ||
| AW109 | 1 [62] | Mononuclear cells |
| ssDNA | 1 [75] | Skeletal muscle |
| Reference | Cells 1 | Groups Compared | Potential Bioactive Compounds (Specifically Indicated in the Article) | Upregulated genes | Downregulated Genes | Main Biological Message Reported in the Article Potentially Associated with the Gene(s) Response |
|---|---|---|---|---|---|---|
| White blood cells | ||||||
| Daak AA et al., 2015 [65] | White blood cells | Omega-3 capsules vs. placebo capsules (high oleic oil blend) | EPA, DHA | - | NFKB1 | Improvement of oxidative stress status and amelioration of inflammation |
| Farràs M et al., 2013 [31] | White blood cells | Olive oil high polyphenols vs. low polyphenols | Mixed olive oil compounds (polyphenols) | ABCA1, SCARB1, PPARG, PPARA, PPARD, MED1, CD36, PTGS1 | - | Enhancement of cholesterol efflux from cells |
| Nieman DC et al., 2007 [80] | White blood cells | Quer vs. placebo | Quer | - | CXCL8, IL10 | Modulation of post-exercise inflammatory status |
| Mononuclear cells | ||||||
| Konstantinidou V et al., 2010 [28] | Mononuclear cells | Med diet + olive oil vs. control diet | Mixed compounds in the Med diet (potential specific contribution of olive oil polyphenols) | - | ADRB2, ARHGAP15, IL7R, POLK, IFNG | Regulation of atherosclerosis-related genes, improvement of oxidative stress and inflammatory status |
| Radler U et al., 2011 [70] | Mononuclear cells | Low-fat yoghurt containing grapeseed extract + fish oil + phospholipids +L- Carn + VitC + VitE (post vs. pre) | Mixed compounds (polyphenols, fatty acids, vitamins) | PPARG, CPT1A, CPT1B, CRAT, SLC22A5 | - | Regulation of fatty acids metabolism |
| Jamilian M et al., 2018 [67] | Mononuclear cells | Fish oil vs. placebo | Mixed compounds (EPA + DHA) | PPARG | LDLR, IL1B, TNF | Improvement of inflammatory status and of insulin and lipid metabolism |
| Plat J & Mensik RP, 2001 [62] | Mononuclear cells | Oils with mixed stanols vs. control (margarine + rapeseed oil) | Mixed compounds (stanol esters: sitos, camp) | LDLR | - | Improvement of LDL-cholesterol metabolism |
| Shrestha S et al., 2007 [57] | Mononuclear cells | Psyllum + plant sterols vs. placebo | Mixed compounds (Psyllum fiber + plant sterols) | LDLR | - | Improvement of LDL-cholesterol metabolism |
| Perez-Herrera A et al., 2013 [32] | Mononuclear cells | Sunflower oil vs. other oils (virgin olive oil, olive antioxidants, mixed oils) | Mixed compounds in sunflower oil | CYBB, NCF1, CYBA, NFE2L2, SOD1, CAT, GSR, GSTP1, TXN | - | Induction of postprandial oxidative stress (potential reduction by oil phenolics) |
| Rangel-Zuñiga OA et al., 2014 [33] | Mononuclear cells | Heated sunflower oil (post vs. pre) | Mixed compounds in sunflower oil | XBP1, HSPA5, CALR | - | Induction of postprandial oxidative stress |
| Camargo A et al., 2010 [27] | Mononuclear cells | Olive oil high polyphenols vs. low polyphenols | Mixed olive oil compounds (polyphenols) | - | EGR1, IL1B | Lessening of deleterious inflammatory profile |
| Castañer O et al., 2012 [30] | Mononuclear cells | Olive oil high polyphenols (Post vs. Pre) or Olive oil high polyphenols vs. low polyphenols | Mixed olive oil compounds (polyphenols) | - | CD40LG, IL23A, IL7R, CXCR2, OLR1, ADRB2, CCL2 | Reduction of atherogenic and inflammatory processes |
| Martín-Peláez S et al ., 2015 [35] | Mononuclear cells | Olive oil high polyphenols (post vs. pre) or Olive oil high polyphenols vs. low polyphenols | Mixed olive oil compounds (polyphenols) | - | ACE, NR1H2, ILR8 | Modulation of the renin-angiotensin-aldosterone system and systolic blood pressure |
| Boss A et al., 2016 [64] | Mononuclear cells | Olive leaf extract vs. placebo (glycerol +sucrose, no polyphenols) | Mixed olive leaf compounds (oleuropein, HTyr) | ID3 | EGR1, PTGS2 | Regulation of inflammatory and lipid metabolism pathways |
| Ghanim H et al., 2010 [58] | Mononuclear cells | Polygonum cuspidatum extract vs. placebo | Mixed compounds in Polygonum cuspidatum extract (Res) | IRS1 | MAPK8, KBKB, PTPN1, SOCS3 | Suppressive effect on oxidative and inflammatory stress |
| Barona J et al., 2012 [50] | Mononuclear cells | Grape powder vs. control | Mixed compounds in grape powder (polyphenols) | NOS2 | - | Anti-oxidative and anti-inflammatory response |
| Tomé-Carneiro J et al., 2013 [51] | Mononuclear cells | Grape extract, Grape extract + Res (post vs. pre or extracts vs. placebo) | Mixed compounds in grape extract (Res) | LRRFIP1 | IL1β, TNF, CCL3, NFKBIA | Beneficial immune-modulatory effect |
| Kropat C et al., 2013 [53] | Mononuclear cells | Bilberry pomace extract (post vs. pre) | Mixed compounds in bilberry pomace extract (anthocyanins) | NQO1 | HMOX1, NFE2L2 | Regulation of antioxidant transcription and antioxidant genes |
| Isolated single type of cells | ||||||
| Persson I et al., 2000 [14] | Lymphocytes | Mix Veg (post vs. pre) | Mixed compounds in the Mix Veg | - | GSTP1 | Compensatory downregulation of endogenous antioxidant systems |
| Hernández-Alonso P et al., 2014 [38] | Lymphocytes | Diet + pistacchio vs. control diet | Mixed compounds in the pistachio (fatty acids, minerals, vitamins, carotenoids, tocopherols polyphenols) | - | IL6, RETN, SLC2A4 | Impact on inflammatory markers of glucose and insulin metabolism |
| Boettler U et al., 2012 [43] | Lymphocytes | Coffee brew (post vs. pre) | Mixed compounds in coffee (CGA, NMP) | NFE2L2 | - | Regulation of antioxidant transcription |
| Volz N et al., 2012 [42] | Lymphocytes | Coffee brew (post vs. pre) | Mixed compounds in coffee (CGA, NMP) | NFE2L2 | HMOX1, SOD1 | Regulation of antioxidant transcription and antioxidant genes |
| Morrow DMP et al., 2001 [79] | Lymphocytes | Quer vs. placebo | Quer | - | TIMP1 | Mediator of carcinogenic processes |
| Marotta F et al., 2010 [45] | Neutrophils | Fermented papaya (Post vs. Pre) | Mixed compounds in fermented papaya | SOD1, CAT, GPX1, OGG1 | - | Regulation of redox balance |
| Carrera-Quintanar L et al., 2015 [60] | Neutrophils | T1: Lippia citriodora extract T2: Almond beverage (+vitC + vitE) T1 + T2 (post vs. pre) | Mixed compounds | - | SOD2, SOD1 | Adaptative antioxidant response |
| Yanaka A et al., 2009 [23] | Polymorpho-nuclear granulocytes | Broccoli sprouts (post vs. pre) | Mixed compounds (SFGluc) | HMOX1 | - | Protective effect against bacterial infection (anti-oxidative, anti-inflammatory) |
| Reference | Cells 1 (n = Number of Samples Analysed) | Groups Compared | Potential Bioactive Compounds (Specifically Indicated in the Article) | Gene Expression Change (FC; % of Change) | Data Quality | Variability (Estimated CV %) | Association with Specific Compounds, Metabolites | Protein Change | Level of Evidence |
|---|---|---|---|---|---|---|---|---|---|
| Inflammation: TNF | |||||||||
| Di Renzo L. et al., 2017 [40] | Blood (n = 22) | McD meal + hazelnuts vs. McD meal (post-) | Mix compounds present in the hazelnuts (fatty acids, polyphenols, etc.) | ↓TNF (FC < -1.5; −34%) | Poor | No information available | No evidence | No evidence | Low |
| Weseler AR et al., 2011 [49] | Blood (n = 15) | Grape seeds (post- vs. pre-) | Mix compounds present in the seeds (flavanols) | ↓TNF (FC = -1.14; −12%) | Poor | No information available | No evidence | Inhibition of ex vivo LPS-induced TNF in blood | Low |
| Tomé-Carneiro J et al., 2013 [51] | Mononuclear cells (n = 9–13) | Grape extract + Res (post- vs. pre-) Grape extract + Res vs. control (post-) | Mix compounds present in the grape extract + Res | ↓TNF (FC = −2.56 to −1.54; −61% to −35%) | High | 3.2–7.9 (only baseline levels) | No evidence | (NC) TNF in serum or plasma | Low |
| Jamilian M et al., 2018 [67] | Mononuclear cells (n = 20) | Fish oil (EPA + DHA) vs. placebo (post-) | Fish oil compounds (EPA + DHA) | ↓TNF (FC = −1.12; −11%) | Medium-Poor | 10.5–18.0 | No evidence | No evidence | Low |
| Vors C. et al., 2017 [72] | Blood (n = 44) | EPA vs. control (post-) DHA vs. control (post-) | EPA, DHA | ↑TNF (EPA, FC = +1.07; 7%)(DHA, FC = +1.09; 9%) | Poor | 19.8–24.2 | No evidence | No correlation between TNF and TNF in plasma | Low |
| Energy metabolism: PPARs | |||||||||
| Radler U et al., 2011 [70] | Mononuclear cells (n = 20–22) | Low-fat yoghurt (grapeseed extract + fish oil + phospholipids + l-carn + vitC + vitE) (post- vs. pre-) | Mix compounds (PUFAs, polyphenols, l-carn) | ↑PPARG (FC = +2.53; 153%) | Poor | 49.8 | No evidence | No evidence | Low |
| Vors C et al., 2017 [72] | Blood (n = 44) | EPA vs. control (post-) DHA vs. control (post-) | EPA DHA | ↑PPARA (EPA, FC = +1.12; 12%) (DHA, FC = +1.10; 10%) | Poor | 17.9–26.1 | No evidence | No evidence | Low |
| Jamilian M et al., 2018 [67] | Mononuclear cells (n = 20) | Fish oil (EPA + DHA) vs. placebo (post-) | Fish oil compounds (EPA + DHA) | ↑PPARG (FC = +1.06; 6%) | Medium-Poor | 9.0–11.3 | No evidence | No evidence | Low |
| Farràs M al., 2013 [31] | White blood cells (n = 13) | High- vs. low-polyphenols in olive oil | Mix olive oil compounds (polyphenols) | ↑PPARG (FC = +2.8; 180%) ↑PPARA (FC = +2.0; 100%) ↑PPARD (FC = +2.0; 100%) ↑MED1 (FC = +1.45; 45%) | Poor | 32.2–118.3 | No evidence | No evidence | Low |
| Antioxidant system: GPXs | |||||||||
| Di Renzo L et al., 2014 [18] | Blood (n = 24) | Red wine Med meal + red wine McD meal + red wine (post- vs. pre-) | Mix compounds in red wine | ↑GPX1 (FC = +1.41; 41%) (FC = +1.52; 52%) (FC = +2.83; 183%) | Poor | No information available | No evidence | No evidence | Low |
| Di Renzo L. et al., 2017 [40] | Blood (n = 22) | McD meal + hazelnuts vs. McD meal (post-) | Mix compounds present in the hazelnuts (fatty acids, polyphenols, etc.) | ↑GPX1, GPX3, GPX4 (F > +1.5; 50%) | Poor | No information available | No evidence | No evidence | Low |
| ↓GPX7 (FC < −1.5; −33%) | |||||||||
| Donadio JLS et al., 2017 [39] | Blood (unclear, n = 130 or n = 12?) | Brazil nuts (with Se) (post- vs. pre-) | Mix compounds present in the brazil nuts (Se, fatty acids, polyphenols, etc.) | ↑GPX1 (FC = +1.3; 30% for a particular genotype) | Poor | No information available | No evidence | No evidence | Low |
| Marotta F et al., 2010 [45] | Neutrophils (n = 11) | Fermented papaya (post- vs. pre-) | Mix compounds present in the fermented papaya | ↑GPX1 (FC = +61–67; >6000% (?)) | Poor | No information available | No evidence | No evidence | Low |
| Reference | Cells 1 | Groups Compared | Potential Bioactive Compounds (Specifically Indicated in the Article) | Upregulated Genes | Downregulated Genes | Genes not Changing or with a Not Significant Change | Association with Metabolites | Effect on Protein Levels | Main biological Message Reported |
|---|---|---|---|---|---|---|---|---|---|
| Olive oil and derived products | |||||||||
| Farràs M et al., 2013 [31] | White blood cells 1 | High vs. moderate polyphenol olive oil (post-) | Olive oil polyphenols | ABCA1, SCARB1, PPARG, PPARA, PPARD, MED1, CD36, PTGS1 | - | (NC) ABCG1, PTGS2 | ↑HTyr acetate in plasma with ↑ABCA1 | NR | Enhancement of cholesterol efflux from cells |
| Konstantinidou V et al., 2010 [28] | Mononuclear cells | Med diet + olive oil (polyphenols) vs control diet (post-) | Mix compounds present in the Med diet and the olive oil (fatty acids, polyphenols, vitamins etc.) | - | ADRB2, ARHGAP15, IL7R, POLK, IFNG | - | ↓IFNG with ↑Tyr in urine (highest dose of olive oil) | ↓ IFNγ in plasma (post- vs. pre-) | Regulation of atherosclerosis-related genes, improvement of oxidative stress and inflammatory status |
| Camargo A et al., 2010 [27] | Mononuclear cells | High vs. low polyphenol olive oil (post-) | Olive oil polyphenols | - | EGR1, IL1B | (NS↓) JUN, PTGS2 | NR | NR | Lessening of deleterious inflammatory profile |
| Castañer O et al., 2012 [30] | Mononuclear cells | High vs. low polyphenol olive oil (post-) | Olive oil polyphenols | - | CD40LG, IL23A, IL7R, CXCR2, OLR1, ADRB2, CCL2 | (NS↓) IFNG, VEGFB, ICAM1 (NC) ALOX5AP, TNFSF10 | ↓OLR1 with the ↑Tyr and ↑HTyr in urine | ↓CCL2 | Reduction of atherogenic and inflammatory processes |
| Hernáez Á et al., 2015 [34] | Mononuclear cells | High vs. low polyphenol olive oil (post-) | Olive oil polyphenols | - | - | (NS↑) LPL | NR | NR | Reduction of LDL concentrations and of LDL atherogenicity |
| Martín-Peláez S et al., 2015 [35] | Mononuclear cells | High vs. low polyphenol olive oil (post-) | Olive oil polyphenols | - | ACE, NR1H2, CXCR2 | (NS↓) CXCR1, ADRB2, MPO, ACE (NC) ECE2, OLR1 | NR | NR | Modulation of the renin-angiotensin-aldosterone system and systolic blood pressure |
| Crespo MC et al., 2015 [63] | Mononuclear cells | Olive mill waste water extract Hytolive (enriched in HTyr) vs. placebo (post-) | Olive waste polyphenols (HTyr) | - | - | (NC) Phase II enzymes: NQO1,2, GSTA1,4, GSTK1, GSTM1-5, GSTO1,2, GSTP1, GSTM1,2, HNMT, INMT, MGST1-3 | NR | NR | Hormesis hypothesis of activation of phase II enzymes by polyphenols |
| Boss A et al., 2016 [64] | Mononuclear cells | Olive leaf extract (oleuropein, HTyr) vs. placebo (post-) | Olive leaf polyphenols (oleuropein, HTyr) | ID3 | EGR1, PTGS2 | - | NR | NR | Regulation of inflammatory and lipid metabolism pathways |
| Kruse M et al., 2015 [36] | Adipose tissue | Olive oil (MUFA) (post- vs. pre-, postpandrial) | Mix olive oil bioactive compounds | CCL2 | - | (NS↓) IL6, IL8 (NS↑) IL10, TNF (NC) IL1β, ADGRE1, SERPINE1 | NR | (NC) MCP-1 (CCL2) | Acute inflammatory and metabolic response related genes |
| Broccoli and derived products | |||||||||
| Atwell LL et al., 2015 [25] | Blood | Broccoli sprout (SFGluc) vs. Myrosinase-treated broccoli sprout extract (SFGluc) (post- vs. pre-) | SFGluc | - | - | (NC) CDKN1A, HMOX1 | NR | (NC) plasma levels of HMOX1 (HO-1) | Search for chemopreventive targets |
| Doss JF et al., 2016 [26] | Blood | Broccoli (SFGluc) | SFGluc | HBG1, HMOX1 | - | (NS↑) NQO1 | NR | (NC) Hbg1 or HbF | Gene expression studies in sickle cell disease (oxidative stress related) |
| Riso P et al., 2010 [24] | Mononuclear cells | Broccoli (SFGluc, Lut, β-car, VitC) (post- vs. pre-) | SFGluc, Lut, β-car, VitC | - | - | (NC) OGG1, NUDT1, HMOX1 | NR | NR | Antioxidant protection related to DNA repairing enzymes |
| Yanaka A et al., 2009 [23] | Polymorpho-nuclear granulocytes | Broccoli sprout (SFGluc) (post- vs. pre-) | SFGluc | HMOX1 | - | - | NR | NR | Protective effect against bacterial infection (anti-oxidative, anti-inflammatory) |
| Gasper AV et al., 2007 [21] | Gastric antrum | Broccoli drink (containing SFGluc) (post- vs. pre-) | SFGluc | GCLM, TXNRD1 | - | (NC) CDKN1A | NR | NR | Effect on xenobiotic metabolism |
| Riedl MA et al., 2009 [22] | Cells from nasal lavage | Broccoli sprout (SFGluc) (different doses) vs. control (alfalfa sprout) | SFGluc | GSTM1, GSTP1, NQO1, HMOX1 | - | - | NR | NR | Effect on Phase II metabolism |
| Grape products and compounds (Res) | |||||||||
| Weseler AR et al., 2011 [49] | Blood | Flavanols isolated from grape seeds (post- vs. pre-) | Flavanols | - | IL6, TNF, IL10 | (NS↓) CAT, GSR, HMOX1 (NC) IL1β, CXCL8, NOS2, NFKBIA, ICAM1, VCAM1, GPX1, GPX4, SOD2 | NR | Plasma: ↓TNF (NC) IL10 | Anti-inflammatory effects in blood |
| Barona J et al., 2012 [50] | Mononuclear cells | Grape powder vs. placebo | Mix compounds in grape (flavonoids) | NOS2 (individuals without dyslipidemia) | - | (NC) CYBB, SOD1, SOD2, GPX1, GPX4 | NR | NR | Anti-oxidative and anti-inflammatory response |
| Tomé-Carneiro J et al., 2013 [51] | Mononuclear cells | Grape extract vs Grape extract + Res (post- vs. pre-) | Polyphenols, Res | LRRFIP1 | IL1β, TNF, CCL3, NFKBIA | (NC) NFKB1 | NR | NC in TNF levels in PBMC or serum | Beneficial immune-modulatory effect |
| Nguyen AV et al., 2009 [48] | Colon tissue (cancer and normal) | Low concentration of grape powder (post- vs. pre-) | Res, flavanols, flavans, anthocyanins, catechin | Normal tissue MYC Cancer tissue MYC, CCND1 | Normal tissue CCND1, AXIN2 | - | NR | NR | Effect on cancer related pathway |
| Mansur AP et al., 2017 [78] | White blood cells | Res (post- vs. pre-; T vs. C) | Res | - | - | (NC) SIRT1 | NR | ↑ Serum hSIRT1 | Comparative study with caloric restriction |
| Chachay VS et al., 2014 [76] | Mononuclear cells | Res (from Polygonium cuspidatum) | Res | - | - | (NC) NQO1, PTP1B, IL6, HMOX1 | NR | ↓plasma IL6 | Effects on non-alcoholic fatty liver disease |
| Yiu EM et al., 2015 [77] | Mononuclear cells | Res (two doses) (post- vs. pre-) | Res | - | - | (NC) FXN (dose 1) (NS↓) FXN (dose 2) | NR | (NC) FXN in PBMC | Effect on the neurodegenerative disease (Friedreich ataxia) |
| Olesen J et al., 2014 [75] | Skeletal muscle | Res (post- vs. pre-; T vs. C) | Res | - | - | (NC) PPARGC1A, TNF, NOS2 | NR | (NC) TNF, iNOS in muscle and plasma | Metabolic and inflammatory status |
| Yoshino J et al., 2012 [73] | Skeletal muscle and adipose tissue | Res (post- vs. pre-; T vs. C) | Res | - | - | (NC) SIRT1, NAMPT, PPARGC1A, UCP3 | NR | NR | Metabolic effects |
| Poulsen MM et al., 2013 [74] | Skeletal muscle and adipose tissue | Res (post- vs. pre-) | Res | - | Muscle SLC2A4 | Muscle (NC) PPARGC1A Adipose (NC) TNF, NFKB1 | NR | NR | Metabolic and inflammatory effects |
| Specific critical issues related to the human gene expression studies revised in this article that need improvement | Strategies to improve the quality of the studies and the level of evidence to support the link between gene response-bioactive compound |
|---|---|
| Human clinical trial design | |
|
|
|
|
|
|
|
|
| qRT-PCR experimental protocols | |
|
|
|
|
|
|
| Data comparison and presentation | |
|
|
|
|
|
|
| Other relevant information needed to prove the relationship between the intake of bioactive compounds and gene expression changes and to increase the level of evidence supporting these molecular changes as a responsive mechanism underlying the health benefits of bioactive compounds in humans | |
|
|
|
|
|
|
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pokimica, B.; García-Conesa, M.-T. Critical Evaluation of Gene Expression Changes in Human Tissues in Response to Supplementation with Dietary Bioactive Compounds: Moving Towards Better-Quality Studies. Nutrients 2018, 10, 807. https://doi.org/10.3390/nu10070807
Pokimica B, García-Conesa M-T. Critical Evaluation of Gene Expression Changes in Human Tissues in Response to Supplementation with Dietary Bioactive Compounds: Moving Towards Better-Quality Studies. Nutrients. 2018; 10(7):807. https://doi.org/10.3390/nu10070807
Chicago/Turabian StylePokimica, Biljana, and María-Teresa García-Conesa. 2018. "Critical Evaluation of Gene Expression Changes in Human Tissues in Response to Supplementation with Dietary Bioactive Compounds: Moving Towards Better-Quality Studies" Nutrients 10, no. 7: 807. https://doi.org/10.3390/nu10070807
APA StylePokimica, B., & García-Conesa, M. -T. (2018). Critical Evaluation of Gene Expression Changes in Human Tissues in Response to Supplementation with Dietary Bioactive Compounds: Moving Towards Better-Quality Studies. Nutrients, 10(7), 807. https://doi.org/10.3390/nu10070807