Effect of Low-Protein Diet and Inulin on Microbiota and Clinical Parameters in Patients with Chronic Kidney Disease
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Design and Subjects
2.2. Patients
2.3. Inclusion Criteria
2.4. Exclusion Criteria
2.5. Laboratory Measurements
2.6. Short Form (SF-36) Health Survey
2.7. NGS Genome Microbiota Data Analysis
3. Results
3.1. Microbial Diversity and Taxonomic Composition Differed between CKD Patients and HC
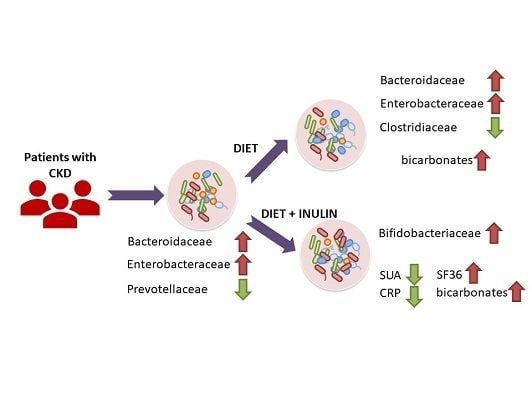
3.2. Effect of Dietary Intervention and Inulin Intake on Gut Microbiota in CKD Patients
3.3. Effect of Dietary Intervention and Inulin Intake on Clincal Parameters in CKD Patients
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Al-Assal, K.; Martinez, A.C.; Torrinhas, R.S.; Cardinelli, C.; Waitzberg, D. Gut microbiota and obesity. Clin. Nutr. Exp. 2018, 20, 60–64. [Google Scholar] [CrossRef] [Green Version]
- Harsch, I.A.; Konturek, P.C. The role of gut microbiota in obesity and type 2 and type 1 diabetes mellitus: New insights into old diseases. Med. Sci. 2018, 6, 32. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Becker, C.; Neurath, M.F.; Wirtz, S. The intestinal microbiota in inflammatory bowel disease. ILAR J. 2015, 56, 192–204. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jie, Z.; Xia, H.; Zhong, S.L.; Feng, Q.; Li, S.; Liang, S.; Zhong, H.; Liu, Z.; Gao, Y.; Zhao, H.; et al. The gut microbiome in atherosclerotic cardiovascular disease. Nat. Commun. 2017, 8, 845. [Google Scholar] [CrossRef] [Green Version]
- Pietrucci, D.; Cerroni, R.; Unida, V.; Farcomeni, A.; Pierantozzi, M.; Mercuri, N.B.; Biocca, S.; Stefani, A.; Desideri, A. Dysbiosis of gut microbiota in a selected population of Parkinson’s patients. Parkinsonism Relat. Disord. 2019, 65, 124–130. [Google Scholar] [CrossRef]
- Ramezani, A.; Raj, D.S. The gut microbiome, kidney disease, and targeted interventions. J. Am. Soc. Nephrol. 2014, 25, 657–670. [Google Scholar] [CrossRef] [Green Version]
- Dou, L.; Bertrand, E.; Cerini, C.; Faure, V.; Sampol, J.; Vanholder, R.; Berland, Y.; Brunet, P. The uremic solutes p-cresol and indoxyl sulfate inhibit endothelial proliferation and wound repair. Kidney Int. 2004, 65, 442–451. [Google Scholar] [CrossRef]
- Vaziri, N.D.; Zhao, Y.Y.; Pahl, M.V. Altered intestinal microbial flora and impaired epithelial barrier structure and function in CKD: The nature, mechanisms, consequences and potential treatment. Nephrol. Dial. Transplant. 2016, 31, 737–746. [Google Scholar] [CrossRef] [Green Version]
- Esgalhado, M.; Kemp, J.A.; Damasceno, N.R.; Fouque, D.; Mafra, D. Short-chain fatty acids: A link between prebiotics and microbiota in chronic kidney disease. Future Microbiol. 2017, 12, 1413–1425. [Google Scholar] [CrossRef]
- Kruse, H.P.; Kleessen, B.; Blaut, M. Effects of inulin on faecal bifidobacteria in human subjects. Br. J. Nutr. 1999, 82, 375–382. [Google Scholar] [CrossRef] [Green Version]
- Levey, A.S.; Coresh, J.; Greene, T.; Stevens, L.A.; Zhang, Y.L.; Hendriksen, S.; Kusek, J.W.; Van Lente, F. Using standardized serum creatinine values in the modification of diet in renal disease study equation for estimating glomerular filtration rate. Ann. Int. Med. 2006, 145, 247–254. [Google Scholar] [CrossRef] [PubMed]
- Lins, L.; Carvalho, F.M. SF-36 total score as a single measure of health-related quality of life: Scoping review. SAGE Open Med. 2016. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, J.; Kobert, K.; Flouri, T.; Stamatakis, A. PEAR: A fast and accurate Illumina Paired-End reAd mergeR. Bioinformatics 2013, 30, 614–620. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Pena, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2012, 41, D590–D596. [Google Scholar] [CrossRef]
- Bokulich, N.A.; Subramanian, S.; Faith, J.J.; Gevers, D.; Gordon, J.I.; Knight, R.; Mills, D.A.; Caporaso, J.G. Quality-filtering vastly improves diversity estimates from Illumina amplicon sequencing. Nat. Methods 2013, 10, 57. [Google Scholar] [CrossRef]
- McMurdie, P.J.; Holmes, S. Waste not, want not: Why rarefying microbiome data is inadmissible. PLoS Comput. Biol. 2014, 10, e1003531. [Google Scholar] [CrossRef] [Green Version]
- Briskey, D.; Tucker, P.; Johnson, D.W.; Coombes, J.S. The role of the gastrointestinal tract and microbiota on uremic toxins and chronic kidney disease development. Clin. Exp. Nephrol. 2017, 21, 7–15. [Google Scholar] [CrossRef]
- Lau, W.L.; Kalantar-Zadeh, K.; Vaziri, N.D. The Gut as a Source of Inflammation in Chronic Kidney Disease. Nephron 2015, 130, 92–98. [Google Scholar] [CrossRef] [Green Version]
- Anders, H.J.; Andersen, K.; Stecher, B. The intestinal microbiota, a leaky gut, and abnormal immunity in kidney disease. Kidney Int. 2013, 83, 1010–1016. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shah, N.B.; Allegretti, A.S.; Nigwekar, S.U.; Kalim, S.; Zhao, S.; Lelouvier, B.; Servant, F.; Serena, G.; Thadhani, R.I.; Raj, D.S.; et al. Blood Microbiome Profile in CKD: A Pilot Study. Clin. J. Am. Soc. Nephrol. 2019, 14, 692–701. [Google Scholar] [CrossRef] [PubMed]
- Sampaio-Maia, B.; Simões-Silva, L.; Pestana, M.; Araujo, R.; Soares-Silva, I.J. The Role of the Gut Microbiome on Chronic Kidney Disease. Adv. Appl. Microbiol. 2016, 96, 65–94. [Google Scholar] [PubMed]
- Li, H.; Li, T.; Beasley, D.E.; Heděnec, P.; Xiao, Z.; Zhang, S.; Li, J.; Lin, Q.; Li, X. Diet Diversity Is Associated with Beta but not Alpha Diversity of Pika Gut Microbiota. Front. Microbiol. 2016, 7, 1169. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, K.; Xia, G.; Lu, J.; Chen, M.; Zhen, X.; Wang, S.; You, C.; Nie, J.; Zhou, H.; Yin, J. Impaired renal function and dysbiosis of gut microbiota contribute to increased trimethylamine-N-oxide in chronic kidney disease patients. Sci. Rep. 2017, 7, 1445. [Google Scholar] [CrossRef]
- Mihai, S.; Codrici, E.; Popescu, I.D.; Enciu, A.M.; Albulescu, L.; Necula, L.G.; Mambet, C.; Anton, G.; Tanase, C. Inflammation-Related Mechanisms in Chronic Kidney Disease Prediction, Progression, and Outcome. J. Immunol. Res. 2018. [Google Scholar] [CrossRef]
- Jazani, N.H.; Savoj, J.; Lustgarten, M.; Lau, W.L.; Vaziri, N.D. Impact of Gut Dysbiosis on Neurohormonal Pathways in Chronic Kidney Disease. Diseases 2019, 7, 21. [Google Scholar] [CrossRef] [Green Version]
- Concistrè, A.; Petramala, L.; Scoccia, G.; Sciomer, S.; Bisogni, V.; Saracino, V.; Iannucci, G.; Lai, S.; Mastroluca, D.; Iacobellis, G.; et al. Epicardial Fat Thickness in Patients with Autosomal Dominant Polycystic Kidney Disease. Cardiorenal. Med. 2018, 8, 199–207. [Google Scholar] [CrossRef]
- Navaneethan, S.D.; Beddhu, S. Associations of serum uric acid with cardiovascular events and mortality in moderate chronic kidney disease. Nephrol. Dial. Transplant. 2009, 24, 1260–1266. [Google Scholar] [CrossRef]
- Lai, S.; Mariotti, A.; Coppola, B.; Lai, C.; Aceto, P.; Dimko, M.; Galani, A.; Innico, G.; Frassetti, N.; Mangiulli, M.; et al. Uricemia and homocysteinemia: Nontraditional risk factors in the early stages of chronic kidney disease—Preliminary data. Eur. Rev. Med. Pharmacol. Sci. 2014, 18, 1010–1017. [Google Scholar]
- Drummond, G.R.; Selemidis, S.; Griendling, K.K.; Sobey, C.G. Combating oxidative stress in vascular disease: NADPH oxidases as therapeutic targets. Nat. Rev. Drug Discov. 2011, 10, 453–471. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Drummond, G.R.; Sobey, C.G. Endothelial NADPH oxidases: Which NOX to target in vascular disease? Trends Endocrinol. Metab. 2014, 25, 452–463. [Google Scholar] [CrossRef] [PubMed]
- Zhao, R.; Ma, X.; Xie, X.; Shen, G.X. Involvement of NADPH oxidase in oxidized LDL-induced upregulation of heat shock factor-1 and plasminogen activator inhibitor-1 in vascular endothelial cells. Am. J. Physiol. Endocrinol. Metab. 2009, 297, E104–E111. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bendall, J.K.; Rinze, R.; Adlam, D.; Tatham, A.L.; de Bono, J.; Wilson, N.; Volpi, E.; Channon, K.M. Endothelial Nox2 overexpression potentiates vascular oxidative stress and hemodynamic response to angiotensin II: Studies in endothelial-targeted Nox2 transgenic mice. Circ. Res. 2007, 100, 1016–1025. [Google Scholar] [CrossRef] [Green Version]
- Douglas, G.; Bendall, J.K.; Crabtree, M.J.; Tatham, A.L.; Carter, E.E.; Hale, A.B.; Channon, K.M. Endothelial-specific Nox2 overexpression increases vascular superoxide and macrophage recruitment in ApoE−/− mice. Cardiovasc. Res. 2012, 94, 20–29. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wesson, D.E.; Mathur, V.; Tangri, N.; Stasiv, Y.; Parsell, D.; Li, E.; Klaerner, G.; Bushinsky, D.A. Long-term safety and efficacy of veverimer in patients with metabolic acidosis in chronic kidney disease: A multicentre, randomised, blinded, placebo-controlled, 40-week extension. Lancet 2019, 394, 396–406. [Google Scholar] [CrossRef]
- Kraut, J.A.; Kurtz, I. Metabolic acidosis of CKD: Diagnosis, clinical characteristics, and treatment. Am. J. Kidney Dis. 2005, 45, 978–993. [Google Scholar] [CrossRef]




© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lai, S.; Molfino, A.; Testorio, M.; Perrotta, A.M.; Currado, A.; Pintus, G.; Pietrucci, D.; Unida, V.; La Rocca, D.; Biocca, S.; et al. Effect of Low-Protein Diet and Inulin on Microbiota and Clinical Parameters in Patients with Chronic Kidney Disease. Nutrients 2019, 11, 3006. https://doi.org/10.3390/nu11123006
Lai S, Molfino A, Testorio M, Perrotta AM, Currado A, Pintus G, Pietrucci D, Unida V, La Rocca D, Biocca S, et al. Effect of Low-Protein Diet and Inulin on Microbiota and Clinical Parameters in Patients with Chronic Kidney Disease. Nutrients. 2019; 11(12):3006. https://doi.org/10.3390/nu11123006
Chicago/Turabian StyleLai, Silvia, Alessio Molfino, Massimo Testorio, Adolfo M. Perrotta, Annachiara Currado, Giovanni Pintus, Daniele Pietrucci, Valeria Unida, Davide La Rocca, Silvia Biocca, and et al. 2019. "Effect of Low-Protein Diet and Inulin on Microbiota and Clinical Parameters in Patients with Chronic Kidney Disease" Nutrients 11, no. 12: 3006. https://doi.org/10.3390/nu11123006
APA StyleLai, S., Molfino, A., Testorio, M., Perrotta, A. M., Currado, A., Pintus, G., Pietrucci, D., Unida, V., La Rocca, D., Biocca, S., & Desideri, A. (2019). Effect of Low-Protein Diet and Inulin on Microbiota and Clinical Parameters in Patients with Chronic Kidney Disease. Nutrients, 11(12), 3006. https://doi.org/10.3390/nu11123006