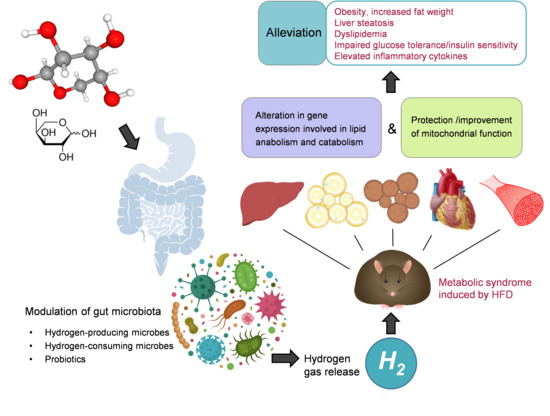
L-Arabinose Elicits Gut-Derived Hydrogen Production and Ameliorates Metabolic Syndrome in C57BL/6J Mice on High-Fat-Diet
Abstract
:1. Introduction
2. Materials and Methods
2.1. Reagents
2.2. Animals and Procedures
2.3. Fecal DNA Extraction
2.4. Histopathologic Analysis
2.5. Measurements of Serum Parameters
2.6. Liver Lipid Profile Measurement
2.7. Quantitative Real-Time PCR
2.8. SDS-PAGE and Western-Blot
2.9. Isolation of Mitochondria and Determination of Mitochondrial Electron-Transport-Chain Complexes Activities
2.10. Statistical Analysis
3. Results
3.1. Oral L-Arabinose Elicits Hydrogen Production in Mice in a Time-and-Dose-Dependent Manner
3.2. L-arabinose Reduces Body Weight, Liver Weight to Body Weight Ratio, Fat Weight to Body Weight Ratio and Serum Leptin Level in Mice on HFD Without Affecting Calorie Intake
3.3. L-Arabinose Improves Glucose Homeostasis in Mice on HFD
3.4. L-arabinose Effectively Alleviates Liver Steatosis As Well As Restores Serum Lipid Profile and Inflammatory Parameters in Mice on HFD
3.5. L-arabinose Modulates mRNA and Protein Expression Pattern of Selected Genes Involved in Lipid Metabolism in Key Lipid-Metabolizing Tissues in Mice on HFD
3.6. L-arabinose Protects Liver and Heart Mitochondrial Function from HFD-Induced Damage and Improves Mitochondrial Function in Muscle in Mice on HFD
3.7. Antibiotics Abolishes Hydrogen Production in Response to L-Arabinose Gavage in Both CD and HFD Fed Mice
3.8. L-arabinose Modulates Relative Abundances of Hydrogen-Producing and Hydrogen-Consuming Gut Microbes in Mice on HFD
3.9. L-arabinose Modulates Relative Abundances of Gut Bacteria Involved in Metabolic Syndrome
4. Discussion
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Hao, L.; Lu, X.; Sun, M.; Li, K.; Shen, L.; Wu, T. Protective effects of L-arabinose in high-carbohydrate, high-fat diet-induced metabolic syndrome in rats. Food Nutr. Res. 2015, 59, 28886. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pan, Y.; Wang, C.; Chen, Z.; Li, W.; Yuan, G.; Chen, H. Physicochemical properties and antidiabetic effects of a polysaccharide from corn silk in high-fat diet and streptozotocin-induced diabetic mice. Carbohydr. Polym. 2017, 164, 370–378. [Google Scholar] [CrossRef] [PubMed]
- Krog-Mikkelsen, I.; Hels, O.; Tetens, I.; Holst, J.J.; Andersen, J.R.; Bukhave, K. The effects of L-arabinose on intestinal sucrase activity: Dose-response studies in vitro and in humans. Am. J. Clin. Nutr. 2011, 94, 472–478. [Google Scholar] [CrossRef] [PubMed]
- Festi, D.; Schiumerini, R.; Eusebi, L.H.; Marasco, G.; Taddia, M.; Colecchia, A. Gut microbiota and metabolic syndrome. World J. Gastroenterol. 2014, 20, 16079–16094. [Google Scholar] [CrossRef] [PubMed]
- Boulange, C.L.; Neves, A.L.; Chilloux, J.; Nicholson, J.K.; Dumas, M.E. Impact of the gut microbiota on inflammation, obesity, and metabolic disease. Genome Med. 2016, 8, 42. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, X.; Devaraj, S. Gut Microbiome in Obesity, Metabolic Syndrome, and Diabetes. Curr. Diab. Rep. 2018, 18, 129. [Google Scholar] [CrossRef] [PubMed]
- Devaraj, S.; Hemarajata, P.; Versalovic, J. The human gut microbiome and body metabolism: Implications for obesity and diabetes. Clin. Chem. 2013, 59, 617–628. [Google Scholar] [CrossRef] [Green Version]
- Murphy, E.A.; Velazquez, K.T.; Herbert, K.M. Influence of high-fat diet on gut microbiota: A driving force for chronic disease risk. Curr. Opin. Clin. Nutr. Metab. Care 2015, 18, 515–520. [Google Scholar] [CrossRef]
- Kim, K.A.; Gu, W.; Lee, I.A.; Joh, E.H.; Kim, D.H. High fat diet-induced gut microbiota exacerbates inflammation and obesity in mice via the TLR4 signaling pathway. PLoS ONE 2012, 7, e47713. [Google Scholar] [CrossRef]
- Lecomte, V.; Kaakoush, N.O.; Maloney, C.A.; Raipuria, M.; Huinao, K.D.; Mitchell, H.M.; Morris, M.J. Changes in gut microbiota in rats fed a high fat diet correlate with obesity-associated metabolic parameters. PLoS ONE 2015, 10, e0126931. [Google Scholar] [CrossRef] [Green Version]
- Marotz, C.A.; Zarrinpar, A. Treating Obesity and Metabolic Syndrome with Fecal Microbiota Transplantation. Yale J. Biol. Med. 2016, 89, 383–388. [Google Scholar] [PubMed]
- Rooks, M.G.; Garrett, W.S. Gut microbiota, metabolites and host immunity. Nat. Rev. Immunol. 2016, 16, 341–352. [Google Scholar] [CrossRef] [PubMed]
- De Vadder, F.; Kovatcheva-Datchary, P.; Goncalves, D.; Vinera, J.; Zitoun, C.; Duchampt, A.; Backhed, F.; Mithieux, G. Microbiota-generated metabolites promote metabolic benefits via gut-brain neural circuits. Cell 2014, 156, 84–96. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sharon, G.; Garg, N.; Debelius, J.; Knight, R.; Dorrestein, P.C.; Mazmanian, S.K. Specialized metabolites from the microbiome in health and disease. Cell Metab. 2014, 20, 719–730. [Google Scholar] [CrossRef] [Green Version]
- Ohta, S. Molecular hydrogen as a preventive and therapeutic medical gas: Initiation, development and potential of hydrogen medicine. Pharmacol. Ther. 2014, 144, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Huang, L. Molecular hydrogen: A therapeutic antioxidant and beyond. Med. Gas Res. 2016, 6, 219–222. [Google Scholar] [CrossRef] [Green Version]
- Ohta, S. Molecular hydrogen as a novel antioxidant: Overview of the advantages of hydrogen for medical applications. Methods Enzymol. 2015, 555, 289–317. [Google Scholar] [CrossRef]
- Hylemon, P.B.; Harris, S.C.; Ridlon, J.M. Metabolism of hydrogen gases and bile acids in the gut microbiome. FEBS Lett. 2018, 592, 2070–2082. [Google Scholar] [CrossRef]
- Rowland, I.; Gibson, G.; Heinken, A.; Scott, K.; Swann, J.; Thiele, I.; Tuohy, K. Gut microbiota functions: Metabolism of nutrients and other food components. Eur. J. Nutr. 2018, 57, 1–24. [Google Scholar] [CrossRef] [Green Version]
- Levitt, M.D. Production and excretion of hydrogen gas in man. N. Engl. J. Med. 1969, 281, 122–127. [Google Scholar] [CrossRef]
- Suzuki, Y.; Sano, M.; Hayashida, K.; Ohsawa, I.; Ohta, S.; Fukuda, K. Are the effects of alpha-glucosidase inhibitors on cardiovascular events related to elevated levels of hydrogen gas in the gastrointestinal tract? FEBS Lett. 2009, 583, 2157–2159. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oku, T.; Nakamura, S. Comparison of digestibility and breath hydrogen gas excretion of fructo-oligosaccharide, galactosyl-sucrose, and isomalto-oligosaccharide in healthy human subjects. Eur. J. Clin. Nutr. 2003, 57, 1150–1156. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wolf, P.G.; Biswas, A.; Morales, S.E.; Greening, C.; Gaskins, H.R. H2 metabolism is widespread and diverse among human colonic microbes. Gut Microbes 2016, 7, 235–245. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ostojic, S.M. Inadequate Production of H2 by Gut Microbiota and Parkinson Disease. Trends Endocrinol. Metab. 2018, 29, 286–288. [Google Scholar] [CrossRef]
- Kamimura, N.; Nishimaki, K.; Ohsawa, I.; Ohta, S. Molecular hydrogen improves obesity and diabetes by inducing hepatic FGF21 and stimulating energy metabolism in db/db mice. Obesity (Silver Spring) 2011, 19, 1396–1403. [Google Scholar] [CrossRef]
- Hou, C.; Wang, Y.; Zhu, E.; Yan, C.; Zhao, L.; Wang, X.; Qiu, Y.; Shen, H.; Sun, X.; Feng, Z.; et al. Coral calcium hydride prevents hepatic steatosis in high fat diet-induced obese rats: A potent mitochondrial nutrient and phase II enzyme inducer. Biochem. Pharmacol. 2016, 103, 85–97. [Google Scholar] [CrossRef]
- Lee, H.K.; Cho, Y.M.; Kwak, S.H.; Lim, S.; Park, K.S.; Shim, E.B. Mitochondrial dysfunction and metabolic syndrome-looking for environmental factors. Biochim. Biophys. Acta 2010, 1800, 282–289. [Google Scholar] [CrossRef]
- Kim, J.A.; Wei, Y.; Sowers, J.R. Role of mitochondrial dysfunction in insulin resistance. Circ. Res. 2008, 102, 401–414. [Google Scholar] [CrossRef]
- Ohta, S. Molecular hydrogen is a novel antioxidant to efficiently reduce oxidative stress with potential for the improvement of mitochondrial diseases. Biochim. Biophys. Acta 2012, 1820, 586–594. [Google Scholar] [CrossRef]
- Ostojic, S.M. Targeting molecular hydrogen to mitochondria: Barriers and gateways. Pharmacol. Res. 2015, 94, 51–53. [Google Scholar] [CrossRef]
- Iuchi, K.; Imoto, A.; Kamimura, N.; Nishimaki, K.; Ichimiya, H.; Yokota, T.; Ohta, S. Molecular hydrogen regulates gene expression by modifying the free radical chain reaction-dependent generation of oxidized phospholipid mediators. Sci. Rep. 2016, 6, 18971. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, X.; Xia, X.; Tang, R.; Zhou, J.; Zhao, H.; Wang, K. Development of a real-time PCR method for Firmicutes and Bacteroidetes in faeces and its application to quantify intestinal population of obese and lean pigs. Lett. Appl. Microbiol. 2008, 47, 367–373. [Google Scholar] [CrossRef] [PubMed]
- Matsuki, T.; Watanabe, K.; Fujimoto, J.; Miyamoto, Y.; Takada, T.; Matsumoto, K.; Oyaizu, H.; Tanaka, R. Development of 16S rRNA-gene-targeted group-specific primers for the detection and identification of predominant bacteria in human feces. Appl. Environ. Microbiol. 2002, 68, 5445–5451. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Balamurugan, R.; Janardhan, H.P.; George, S.; Raghava, M.V.; Muliyil, J.; Ramakrishna, B.S. Molecular studies of fecal anaerobic commensal bacteria in acute diarrhea in children. J. Pediatr. Gastroenterol. Nutr. 2008, 46, 514–519. [Google Scholar] [CrossRef] [PubMed]
- Ramirez-Farias, C.; Slezak, K.; Fuller, Z.; Duncan, A.; Holtrop, G.; Louis, P. Effect of inulin on the human gut microbiota: Stimulation of Bifidobacterium adolescentis and Faecalibacterium prausnitzii. Br. J. Nutr. 2009, 101, 541–550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dridi, B.; Henry, M.; El Khechine, A.; Raoult, D.; Drancourt, M. High prevalence of Methanobrevibacter smithii and Methanosphaera stadtmanae detected in the human gut using an improved DNA detection protocol. PLoS ONE 2009, 4, e7063. [Google Scholar] [CrossRef] [PubMed]
- Fite, A.; Macfarlane, G.T.; Cummings, J.H.; Hopkins, M.J.; Kong, S.C.; Furrie, E.; Macfarlane, S. Identification and quantitation of mucosal and faecal desulfovibrios using real time polymerase chain reaction. Gut 2004, 53, 523–529. [Google Scholar] [CrossRef] [Green Version]
- Delroisse, J.M.; Boulvin, A.L.; Parmentier, I.; Dauphin, R.D.; Vandenbol, M.; Portetelle, D. Quantification of Bifidobacterium spp. and Lactobacillus spp. in rat fecal samples by real-time PCR. Microbiol. Res. 2008, 163, 663–670. [Google Scholar] [CrossRef]
- Larsen, N.; Vogensen, F.K.; van den Berg, F.W.; Nielsen, D.S.; Andreasen, A.S.; Pedersen, B.K.; Al-Soud, W.A.; Sorensen, S.J.; Hansen, L.H.; Jakobsen, M. Gut microbiota in human adults with type 2 diabetes differs from non-diabetic adults. PLoS ONE 2010, 5, e9085. [Google Scholar] [CrossRef]
- Nadkarni, M.A.; Martin, F.E.; Jacques, N.A.; Hunter, N. Determination of bacterial load by real-time PCR using a broad-range (universal) probe and primers set. Microbiology 2002, 148, 257–266. [Google Scholar] [CrossRef] [Green Version]
- Sun, L.; Shen, W.; Liu, Z.; Guan, S.; Liu, J.; Ding, S. Endurance exercise causes mitochondrial and oxidative stress in rat liver: Effects of a combination of mitochondrial targeting nutrients. Life Sci. 2010, 86, 39–44. [Google Scholar] [CrossRef] [PubMed]
- Reddy, P.; Lent-Schochet, D.; Ramakrishnan, N.; McLaughlin, M.; Jialal, I. Metabolic syndrome is an inflammatory disorder: A conspiracy between adipose tissue and phagocytes. Clin. Chim. Acta 2019, 496, 35–44. [Google Scholar] [CrossRef] [PubMed]
- Oh, D.Y.; Talukdar, S.; Bae, E.J.; Imamura, T.; Morinaga, H.; Fan, W.; Li, P.; Lu, W.J.; Watkins, S.M.; Olefsky, J.M. GPR120 is an omega-3 fatty acid receptor mediating potent anti-inflammatory and insulin-sensitizing effects. Cell 2010, 142, 687–698. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheng, Z.; Tseng, Y.; White, M.F. Insulin signaling meets mitochondria in metabolism. Trends Endocrinol. Metab. 2010, 21, 589–598. [Google Scholar] [CrossRef] [Green Version]
- Newberry, C.; Tierney, A.; Pickett-Blakely, O. Lactulose Hydrogen Breath Test Result Is Associated with Age and Gender. Biomed. Res. Int. 2016, 2016, 1064029. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ley, R.E.; Turnbaugh, P.J.; Klein, S.; Gordon, J.I. Microbial ecology: Human gut microbes associated with obesity. Nature 2006, 444, 1022–1023. [Google Scholar] [CrossRef]
- Bell, D.S. Changes seen in gut bacteria content and distribution with obesity: Causation or association? Postgrad. Med. 2015, 127, 863–868. [Google Scholar] [CrossRef]
- Kobyliak, N.; Conte, C.; Cammarota, G.; Haley, A.P.; Styriak, I.; Gaspar, L.; Fusek, J.; Rodrigo, L.; Kruzliak, P. Probiotics in prevention and treatment of obesity: A critical view. Nutr. Metab. (Lond.) 2016, 13, 14. [Google Scholar] [CrossRef] [Green Version]
- Kajiyama, S.; Hasegawa, G.; Asano, M.; Hosoda, H.; Fukui, M.; Nakamura, N.; Kitawaki, J.; Imai, S.; Nakano, K.; Ohta, M.; et al. Supplementation of hydrogen-rich water improves lipid and glucose metabolism in patients with type 2 diabetes or impaired glucose tolerance. Nutr. Res. 2008, 28, 137–143. [Google Scholar] [CrossRef]
- Harris, S.C.; Devendran, S.; Mendez-Garcia, C.; Mythen, S.M.; Wright, C.L.; Fields, C.J.; Hernandez, A.G.; Cann, I.; Hylemon, P.B.; Ridlon, J.M. Bile acid oxidation by Eggerthella lenta strains C592 and DSM 2243(T). Gut Microbes 2018, 9, 523–539. [Google Scholar] [CrossRef] [Green Version]
- Sun, L.; Xie, C.; Wang, G.; Wu, Y.; Wu, Q.; Wang, X.; Liu, J.; Deng, Y.; Xia, J.; Chen, B.; et al. Gut microbiota and intestinal FXR mediate the clinical benefits of metformin. Nat. Med. 2018, 24, 1919–1929. [Google Scholar] [CrossRef] [PubMed]
- Islam, K.B.; Fukiya, S.; Hagio, M.; Fujii, N.; Ishizuka, S.; Ooka, T.; Ogura, Y.; Hayashi, T.; Yokota, A. Bile acid is a host factor that regulates the composition of the cecal microbiota in rats. Gastroenterology 2011, 141, 1773–1781. [Google Scholar] [CrossRef] [PubMed]
- Wahlstrom, A.; Sayin, S.I.; Marschall, H.U.; Backhed, F. Intestinal Crosstalk between Bile Acids and Microbiota and Its Impact on Host Metabolism. Cell Metab. 2016, 24, 41–50. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ostojic, S.M. Non-gut microbiota as a source of bioactive hydrogen. Postgrad. Med. J. 2017, 93, 170. [Google Scholar] [CrossRef]
- Strocchi, A.; Levitt, M.D. Factors affecting hydrogen production and consumption by human fecal flora. The critical roles of hydrogen tension and methanogenesis. J. Clin. Investig. 1992, 89, 1304–1311. [Google Scholar] [CrossRef]
- Triantafyllou, K.; Chang, C.; Pimentel, M. Methanogens, methane and gastrointestinal motility. J. Neurogastroenterol. Motil. 2014, 20, 31–40. [Google Scholar] [CrossRef] [Green Version]
- Mathur, R.; Kim, G.; Morales, W.; Sung, J.; Rooks, E.; Pokkunuri, V.; Weitsman, S.; Barlow, G.M.; Chang, C.; Pimentel, M. Intestinal Methanobrevibacter smithii but not total bacteria is related to diet-induced weight gain in rats. Obesity (Silver Spring) 2013, 21, 748–754. [Google Scholar] [CrossRef]
- Ishibashi, T. Molecular hydrogen: New antioxidant and anti-inflammatory therapy for rheumatoid arthritis and related diseases. Curr. Pharm. Des. 2013, 19, 6375–6381. [Google Scholar] [CrossRef] [Green Version]
- Ragsdale, S.W.; Pierce, E. Acetogenesis and the Wood-Ljungdahl pathway of CO(2) fixation. Biochim. Biophys. Acta 2008, 1784, 1873–1898. [Google Scholar] [CrossRef] [Green Version]
- Lin, S.; Thomas, T.C.; Storlien, L.H.; Huang, X.F. Development of high fat diet-induced obesity and leptin resistance in C57Bl/6J mice. Int. J. Obes. Relat. Metab. Disord. 2000, 24, 639–646. [Google Scholar] [CrossRef] [Green Version]
- Druce, M.; Bloom, S.R. The regulation of appetite. Arch. Dis. Child. 2006, 91, 183–187. [Google Scholar] [CrossRef] [PubMed]
- Perry, B.; Wang, Y. Appetite regulation and weight control: The role of gut hormones. Nutr. Diabetes 2012, 2, e26. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Neary, N.M.; Goldstone, A.P.; Bloom, S.R. Appetite regulation: From the gut to the hypothalamus. Clin. Endocrinol. (Oxf.) 2004, 60, 153–160. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, H.; Storlien, L.H.; Huang, X.F. Effects of dietary fat types on body fatness, leptin, and ARC leptin receptor, NPY, and AgRP mRNA expression. Am. J. Physiol. Endocrinol. Metab. 2002, 282, E1352–E1359. [Google Scholar] [CrossRef] [Green Version]
- Choi, M.S.; Kim, Y.J.; Kwon, E.Y.; Ryoo, J.Y.; Kim, S.R.; Jung, U.J. High-fat diet decreases energy expenditure and expression of genes controlling lipid metabolism, mitochondrial function and skeletal system development in the adipose tissue, along with increased expression of extracellular matrix remodelling- and inflammation-related genes. Br. J. Nutr. 2015, 113, 867–877. [Google Scholar] [CrossRef] [Green Version]









| N/A | Chow Diet | High Fat Diet | ||||
|---|---|---|---|---|---|---|
| Ingredient | gm | kcal | Calorie% | gm | kcal | Calorie% |
| Casein, 30 Mesh | 200 | 800 | 19.72 | 200 | 800 | 19.72 |
| L-Cystine | 3 | 12 | 0.30 | 3 | 12 | 0.30 |
| Corn Starch | 506.3 | 2024.8 | 49.91 | 0 | 0 | 0 |
| Maltodextrin 10 | 125 | 500 | 12.32 | 125 | 500 | 12.32 |
| Sucrose | 68.8 | 275.2 | 6.78 | 68.8 | 275 | 6.78 |
| Cellulose, BW200 | 50 | 0 | 0 | 50 | 0 | 0 |
| Soybean Oil | 25 | 225 | 5.55 | 25 | 225 | 5.55 |
| Lard | 20 | 180 | 4.44 | 245 | 2205 | 54.35 |
| Mineral Mix S10026 | 10 | 0 | 0 | 10 | 0 | 0 |
| Dicalcium Phosphate | 13 | 0 | 0 | 13 | 0 | 0 |
| Calcium Carbonate | 5.5 | 0 | 0 | 5.5 | 0 | 0 |
| Potassium Citrate, 1 H2O | 16.5 | 0 | 0 | 16.5 | 0 | 0 |
| Vitamin Mix V10001 | 10 | 40 | 0.99 | 10 | 40 | 0.99 |
| Choline Bitartrate | 2 | 0 | 0 | 2 | 0 | 0 |
| FD & C Red Dye #40 | 0.04 | 0 | 0 | 0 | 0 | 0 |
| FD & C Blue Dye #1 | 0.01 | 0 | 0 | 0.05 | 0 | 0 |
| Total | 1055.05 | 4057 | 100 | 773.85 | 4057 | 100 |
| Component | gm% | Calorie% | gm% | Calorie% | ||
| Protein | 19.2 | 20 | 26 | 20 | ||
| Carbohydrate | 67.3 | 70 | 26 | 20 | ||
| Fat | 4.3 | 10 | 36 | 60 | ||
| kcal/gm | 3.85 | 5.24 | ||||
| Abbreviation | Full Gene Name | Forward/Reverse Primer Sequence |
|---|---|---|
| Pparα | peroxisome proliferator-activated receptor alpha | F: CAAGGCCTCAGGGTACCACTAC |
| R: GCCGAATAGTTCGCCGAAA | ||
| Pparγ | peroxisome proliferator-activated receptor gamma | F: ATTGAGTGCCGAGTCTGTGG |
| R: ACCTGATGGCATTGTGAGACA | ||
| Srebf1 | sterol regulatory element binding transcription factor 1 | F: CAGACTCACTGCTGCTGACA |
| R: GATGGTCCCTCCACTCACCA | ||
| Acadm | acyl-Coenzyme A dehydrogenase, medium chain | F: GGAGTACCCGTTCCCTCTCA |
| R: CCATACGCCAACTCTTCGGT | ||
| Hmgcr | 3-hydroxy-3-methylglutaryl-Coenzyme A reductase | F: CTTGTGGAATGCCTTGTGATTG |
| R: AGCCGAAGCAGCACATGAT | ||
| Ffar4 | free fatty acid receptor 4 | F: ACCAAGTCAATCGCACCCAC |
| R: GTGAGACGACAAAGATGAGCC | ||
| Fasn | fatty acid synthase | F: GGCCCCTCTGTTAATTGGCT |
| R: CGCTTGTTGGTGGACACTTG | ||
| Elovl3 | ELOVLa fatty acid elongase 3 | F: ATGAATTTCTCACGCGGGTT |
| R: TGTAGGTCTGGCCAACAACG | ||
| Elovl6 | ELOVL fatty acid elongase 6 | F: TCTGATGAACAAGCGAGCCA |
| R: TGAAGACGGCAAGAGTCAGC | ||
| Cpt1 | carnitine palmitoyl transferase 1 | F: CATCCACGCCATACTGCT |
| R: GACCTTGAAGTAACGGCCTC | ||
| Cpt2 | carnitine palmitoyl transferase 2 | F: CAGAGACAGCACTCAGACCC |
| R: TTCTCCTTAGCAGCGGTGAC | ||
| Ucp1 | uncoupling protein 1 | F: CACGGGGACCTACAATGCTT |
| R: ACAGTAAATGGCAGGGGACG | ||
| Ucp3 | uncoupling protein 3 | F: GATACGCCTGGGAACTGGAG |
| R: GGAGCGTTCATGTATCGGGT | ||
| Leptin | Leptin | F: AGGATGACACCAAAACCCTC |
| R: TCTTGGACAAACTCAGAATGGG | ||
| Ap2/Fabp4 | adipocyte protein 2/fatty acid binding protein 4 | F: TGGAAGCTTGTCTCCAGTGA |
| R: AATCCCCATTTACGCTGATG | ||
| Lpl | lipoprotein lipase | F: TTCAACCACAGCAGCAAGAC |
| R: TTCTCTCTTGTACAGGGCGG | ||
| Acox1 | acyl-Coenzyme A oxidase 1 | F: GGGAATTTGGCATCGCAGAC |
| R: ATTGAGGCCAACAGGTTCCA | ||
| Gapdh | Glyceraldehyde 3-phosphate dehydrogenase | F: AGGTCGGTGTGAACGGATTTG |
| R: TGTAGACCATGTAGTTGAGGTCA |
| Gut Microbes | Forward Primer Sequence | Reverse Primer Sequence | Reference |
|---|---|---|---|
| Bacteroidetes | GGARCATGTGGTTTAATTCGATGAT | AGCTGACGACAACCATGCAG | [32] |
| Firmicutes | GGAGYATGTGGTTTAATTCGAAGCA | AGCTGACGACAACCATGCAC | [32] |
| Clostridium coccoides group | AAATGACGGTACCTGACTAA | CTTTGAGTTTCATTCTTGCGAA | [33] |
| Eubacterium rectale | AAGGGAAGCAACGCTGTGAA | CGGTTAGGTCACTGGCTTC | [34] |
| Cluster Ⅳ Ruminococcus spp. | GGCGGCYTRCTGGGCTTT | CCAGGTGGATWACTTATTGTGTTAA | [35] |
| Anaerostipes caccae | TAGCCAGCATTTGAGGTGGG | CTCACGACTTCGCTTCCCTT | N/A |
| Victivallis vadensis | TAAGTTGACCGCCTGGGAAC | CCAGGTAAGGTTCTTCGCGT | N/A |
| Methanobrevibacter smithii | CCGGGTATCTAATCCGGTTC | CTCCCAGGGTAGAGGTGAAA | [36] |
| Desulfovibrios | CCGTAGATATCTGGAGGAACATCAG | ACATCTAGCATCCATCGTTTACAGC | [37] |
| Bifidobacterium spp. | CGCGTCYGGTGTGAAAG | CCCCACATCCAGCATCCA | [38] |
| Lactobacillus group | AGCAGTAGGGAATCTTCCA | CACCGCTACACATGGAG | [39] |
| Domain bacteria universal | TCCTACGGGAGGCAGCAGT | GGACTACCAGGGTATCTAATCCTGTT | [40] |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, L.; Wang, Y.; Zhang, G.; Zhang, T.; Lou, J.; Liu, J. L-Arabinose Elicits Gut-Derived Hydrogen Production and Ameliorates Metabolic Syndrome in C57BL/6J Mice on High-Fat-Diet. Nutrients 2019, 11, 3054. https://doi.org/10.3390/nu11123054
Zhao L, Wang Y, Zhang G, Zhang T, Lou J, Liu J. L-Arabinose Elicits Gut-Derived Hydrogen Production and Ameliorates Metabolic Syndrome in C57BL/6J Mice on High-Fat-Diet. Nutrients. 2019; 11(12):3054. https://doi.org/10.3390/nu11123054
Chicago/Turabian StyleZhao, Lin, Yan Wang, Guanfei Zhang, Tiantian Zhang, Jing Lou, and Jiankang Liu. 2019. "L-Arabinose Elicits Gut-Derived Hydrogen Production and Ameliorates Metabolic Syndrome in C57BL/6J Mice on High-Fat-Diet" Nutrients 11, no. 12: 3054. https://doi.org/10.3390/nu11123054
APA StyleZhao, L., Wang, Y., Zhang, G., Zhang, T., Lou, J., & Liu, J. (2019). L-Arabinose Elicits Gut-Derived Hydrogen Production and Ameliorates Metabolic Syndrome in C57BL/6J Mice on High-Fat-Diet. Nutrients, 11(12), 3054. https://doi.org/10.3390/nu11123054