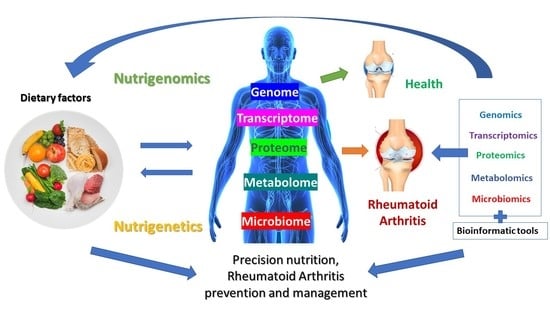
Nutrition and Rheumatoid Arthritis in the ‘Omics’ Era
Abstract
:1. Introduction
2. Applications of ‘Omics’ Approaches and Technologies within Nutritional Research
3. The Contribution of ‘Omics’ in Elucidating Rheumatoid Arthritis Pathogenesis
4. Nutrigenomics Approach to Rheumatoid Arthritis
5. Rheumatoid Arthritis, Microbiome and Nutrition
6. Discussion
7. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Donlin, L.T.; Park, S.-H.; Giannopoulou, E.; Ivovic, A.; Park-Min, K.-H.; Siegel, R.M.; Ivashkiv, L.B. Insights into rheumatic diseases from next-generation sequencing. Nat. Rev. Rheumatol. 2019, 15, 327–339. [Google Scholar] [CrossRef] [PubMed]
- Nemtsova, M.V.; Zaletaev, D.V.; Bure, I.V.; Mikhaylenko, D.S.; Kuznetsova, E.B.; Alekseeva, E.A.; Beloukhova, M.I.; Deviatkin, A.A.; Lukashev, A.N.; Zamyatnin, A.A. Epigenetic Changes in the Pathogenesis of Rheumatoid Arthritis. Front. Genet. 2019, 10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kishikawa, T.; Maeda, Y.; Nii, T.; Motooka, D.; Matsumoto, Y.; Matsushita, M.; Matsuoka, H.; Yoshimura, M.; Kawada, S.; Teshigawara, S.; et al. Metagenome-wide association study of gut microbiome revealed novel aetiology of rheumatoid arthritis in the Japanese population. Ann. Rheum. Dis. 2020, 79, 103. [Google Scholar] [CrossRef] [PubMed]
- Sales, N.M.R.; Pelegrini, P.B.; Goersch, M.C. Nutrigenomics: Definitions and advances of this new science. J. Nutr. Metab. 2014, 2014, 202759. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gioia, C.; Lucchino, B.; Tarsitano, M.G.; Iannuccelli, C.; Di Franco, M. Dietary Habits and Nutrition in Rheumatoid Arthritis: Can Diet Influence Disease Development and Clinical Manifestations? Nutrients 2020, 12, 1456. [Google Scholar] [CrossRef] [PubMed]
- He, J.; Wang, Y.; Feng, M.; Zhang, X.; Jin, Y.-B.; Li, X.; Su, L.-C.; Liu, S.; Wang, A.-X.; Chen, X.-M.; et al. Dietary intake and risk of rheumatoid arthritis-a cross section multicenter study. Clin. Rheumatol. 2016, 35, 2901–2908. [Google Scholar] [CrossRef]
- Maeda, Y.; Takeda, K. Host–microbiota interactions in rheumatoid arthritis. Exp. Mol. Med. 2019, 51, 1–6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Horta-Baas, G.; Romero-Figueroa, M.d.S.; Montiel-Jarquín, A.J.; Pizano-Zárate, M.L.; García-Mena, J.; Ramírez-Durán, N. Intestinal Dysbiosis and Rheumatoid Arthritis: A Link between Gut Microbiota and the Pathogenesis of Rheumatoid Arthritis. J. Immunol. Res. 2017, 2017, 4835189. [Google Scholar] [CrossRef] [PubMed]
- Sela, D.A.; Mills, D.A. The marriage of nutrigenomics with the microbiome: The case of infant-associated bifidobacteria and milk. Am. J. Clin. Nutr. 2014, 99, 697S–703S. [Google Scholar] [CrossRef] [Green Version]
- Di Bella, J.M.; Bao, Y.; Gloor, G.B.; Burton, J.P.; Reid, G. High throughput sequencing methods and analysis for microbiome research. J. Microbiol. Methods 2013, 95, 401–414. [Google Scholar] [CrossRef]
- Lloyd-Price, J.; Arze, C.; Ananthakrishnan, A.N.; Schirmer, M.; Avila-Pacheco, J.; Poon, T.W.; Andrews, E.; Ajami, N.J.; Bonham, K.S.; Brislawn, C.J.; et al. Multi-omics of the gut microbial ecosystem in inflammatory bowel diseases. Nature 2019, 569, 655–662. [Google Scholar] [CrossRef] [PubMed]
- Dei-Cas, I.; Giliberto, F.; Luce, L.; Dopazo, H.; Penas-Steinhardt, A. Metagenomic analysis of gut microbiota in non-treated plaque psoriasis patients stratified by disease severity: Development of a new Psoriasis-Microbiome Index. Sci. Rep. 2020, 10, 12754. [Google Scholar] [CrossRef] [PubMed]
- Eetemadi, A.; Rai, N.; Pereira, B.M.P.; Kim, M.; Schmitz, H.; Tagkopoulos, I. The Computational Diet: A Review of Computational Methods Across Diet, Microbiome, and Health. Front. Microbiol. 2020, 11, 393. [Google Scholar] [CrossRef] [Green Version]
- Leeming, E.R.; Johnson, A.J.; Spector, T.D.; Le Roy, C.I. Effect of Diet on the Gut Microbiota: Rethinking Intervention Duration. Nutrients 2019, 11, 2862. [Google Scholar] [CrossRef] [Green Version]
- Kolodziejczyk, A.A.; Zheng, D.; Elinav, E. Diet–microbiota interactions and personalized nutrition. Nat. Rev. Microbiol. 2019, 17, 742–753. [Google Scholar] [CrossRef]
- Nasir, A.; Bullo, M.M.H.; Ahmed, Z.; Imtiaz, A.; Yaqoob, E.; Jadoon, M.; Ahmed, H.; Afreen, A.; Yaqoob, S. Nutrigenomics: Epigenetics and cancer prevention: A comprehensive review. Crit. Rev. Food Sci. Nutr. 2020, 60, 1375–1387. [Google Scholar] [CrossRef]
- De Angelis, M.; Ferrocino, I.; Calabrese, F.M.; De Filippis, F.; Cavallo, N.; Siragusa, S.; Rampelli, S.; Di Cagno, R.; Rantsiou, K.; Vannini, L.; et al. Diet influences the functions of the human intestinal microbiome. Sci. Rep. 2020, 10, 4247. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yoshida, N.; Yamashita, T.; Hirata, K.-I. Gut Microbiome and Cardiovascular Diseases. Diseases 2018, 6, 56. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ferguson Jane, F.; Allayee, H.; Gerszten Robert, E.; Ideraabdullah, F.; Kris-Etherton Penny, M.; Ordovás José, M.; Rimm Eric, B.; Wang Thomas, J.; Bennett Brian, J. Nutrigenomics, the Microbiome, and Gene-Environment Interactions: New Directions in Cardiovascular Disease Research, Prevention, and Treatment. Circ. Cardiovasc. Genet. 2016, 9, 291–313. [Google Scholar] [CrossRef] [Green Version]
- Dongiovanni, P.; Valenti, L. A Nutrigenomic Approach to Non-Alcoholic Fatty Liver Disease. Int. J. Mol. Sci. 2017, 18, 1534. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Al Theyab, A.; Almutairi, T.; Al-Suwaidi, A.M.; Bendriss, G.; McVeigh, C.; Chaari, A. Epigenetic Effects of Gut Metabolites: Exploring the Path of Dietary Prevention of Type 1 Diabetes. Front. Nutr. 2020, 7. [Google Scholar] [CrossRef]
- Ferguson, L.R. Nutrigenetics, nutrigenomics and inflammatory bowel diseases. Expert Rev. Clin. Immunol. 2013, 9, 717–726. [Google Scholar] [CrossRef] [PubMed]
- Berná, G.; Oliveras-López, M.J.; Jurado-Ruíz, E.; Tejedo, J.; Bedoya, F.; Soria, B.; Martín, F. Nutrigenetics and nutrigenomics insights into diabetes etiopathogenesis. Nutrients 2014, 6, 5338–5369. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ren, X.; Li, X. Advances in Research on Diabetes by Human Nutriomics. Int. J. Mol. Sci. 2019, 20, 5375. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Taylor, P.C.; Moore, A.; Vasilescu, R.; Alvir, J.; Tarallo, M. A structured literature review of the burden of illness and unmet needs in patients with rheumatoid arthritis: A current perspective. Rheumatol. Int. 2016, 36, 685–695. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sami, W.; Ansari, T.; Butt, N.S.; Hamid, M.R.A. Effect of diet on type 2 diabetes mellitus: A review. Int. J. Health Sci. (Qassim) 2017, 11, 65–71. [Google Scholar]
- Popkin, B.M. Nutrition Transition and the Global Diabetes Epidemic. Curr. Diabetes Rep. 2015, 15, 64. [Google Scholar] [CrossRef] [PubMed]
- Casas, R.; Castro-Barquero, S.; Estruch, R.; Sacanella, E. Nutrition and Cardiovascular Health. Int. J. Mol. Sci. 2018, 19, 3988. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Becerra-Tomás, N.; Paz-Graniel, I.; WC Kendall, C.; Kahleova, H.; Rahelić, D.; Sievenpiper, J.L.; Salas-Salvadó, J. Nut consumption and incidence of cardiovascular diseases and cardiovascular disease mortality: A meta-analysis of prospective cohort studies. Nutr. Rev. 2019, 77, 691–709. [Google Scholar] [CrossRef]
- Marklund, M.; Wu, J.H.Y.; Imamura, F.; Del Gobbo, L.C.; Fretts, A.; de Goede, J.; Shi, P.; Tintle, N.; Wennberg, M.; Aslibekyan, S.; et al. Biomarkers of Dietary Omega-6 Fatty Acids and Incident Cardiovascular Disease and Mortality. Circulation 2019, 139, 2422–2436. [Google Scholar] [CrossRef] [Green Version]
- Shirlina, N.G.; Vilms, E.A.; Stasenko, V.L. Nutrition as a possible risk factor for breast cancer. Gig. Sanit. 2015, 94, 48–52. [Google Scholar] [PubMed]
- Thanikachalam, K.; Khan, G. Colorectal Cancer and Nutrition. Nutrients 2019, 11, 164. [Google Scholar] [CrossRef] [Green Version]
- Key, T.J.; Bradbury, K.E.; Perez-Cornago, A.; Sinha, R.; Tsilidis, K.K.; Tsugane, S. Diet, nutrition, and cancer risk: What do we know and what is the way forward? BMJ 2020, 368, m511. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gonzales, J.F.; Barnard, N.D.; Jenkins, D.J.; Lanou, A.J.; Davis, B.; Saxe, G.; Levin, S. Applying the precautionary principle to nutrition and cancer. J. Am. Coll. Nutr. 2014, 33, 239–246. [Google Scholar] [CrossRef]
- Pistollato, F.; Iglesias, R.C.; Ruiz, R.; Aparicio, S.; Crespo, J.; Lopez, L.D.; Manna, P.P.; Giampieri, F.; Battino, M. Nutritional patterns associated with the maintenance of neurocognitive functions and the risk of dementia and Alzheimer’s disease: A focus on human studies. Pharmacol. Res. 2018, 131, 32–43. [Google Scholar] [CrossRef] [PubMed]
- Seidl, S.E.; Santiago, J.A.; Bilyk, H.; Potashkin, J.A. The emerging role of nutrition in Parkinson’s disease. Front. Aging Neurosci. 2014, 6, 36. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Johansson, K.; Askling, J.; Alfredsson, L.; Di Giuseppe, D.; on behalf of the EIRA Study Group. Mediterranean diet and risk of rheumatoid arthritis: A population-based case-control study. Arthritis Res. Ther. 2018, 20, 175. [Google Scholar] [CrossRef]
- Skoczyńska, M.; Świerkot, J. The role of diet in rheumatoid arthritis. Reumatologia 2018, 56, 259–267. [Google Scholar] [CrossRef] [PubMed]
- Valdés, A.; Cifuentes, A.; León, C. Foodomics evaluation of bioactive compounds in foods. TrAC Trends Anal. Chem. 2017, 96, 2–13. [Google Scholar] [CrossRef]
- Aruoma, O.I.; Hausman-Cohen, S.; Pizano, J.; Schmidt, M.A.; Minich, D.M.; Joffe, Y.; Brandhorst, S.; Evans, S.J.; Brady, D.M. Personalized Nutrition: Translating the Science of Nutrigenomics into Practice: Proceedings from the 2018 American College of Nutrition Meeting. J. Am. Coll. Nutr. 2019, 38, 287–301. [Google Scholar] [CrossRef] [PubMed]
- Trujillo, E.; Davis, C.; Milner, J. Nutrigenomics, proteomics, metabolomics, and the practice of dietetics. J. Am. Diet. Assoc. 2006, 106, 403–413. [Google Scholar] [CrossRef]
- El-Sohemy, A. Nutrigenetics. Forum Nutr. 2007, 60, 25–30. [Google Scholar] [CrossRef] [PubMed]
- Gomez-Delgado, F.; Alcala-Diaz, J.F.; Garcia-Rios, A.; Delgado-Lista, J.; Ortiz-Morales, A.; Rangel-Zuñiga, O.; Tinahones, F.J.; Gonzalez-Guardia, L.; Malagon, M.M.; Bellido-Muñoz, E.; et al. Polymorphism at the TNF-alpha gene interacts with Mediterranean diet to influence triglyceride metabolism and inflammation status in metabolic syndrome patients: From the CORDIOPREV clinical trial. Mol. Nutr. Food Res. 2014, 58, 1519–1527. [Google Scholar] [CrossRef]
- Cornelis, M.C.; El-Sohemy, A.; Kabagambe, E.K.; Campos, H. Coffee, CYP1A2 genotype, and risk of myocardial infarction. JAMA 2006, 295, 1135–1141. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ordovas, J.M.; Corella, D. Nutritional genomics. Annu. Rev. Genom. Hum. Genet. 2004, 5, 71–118. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cornelis, M.C.; El-Sohemy, A. Coffee, caffeine, and coronary heart disease. Curr. Opin. Lipidol. 2007, 18, 13–19. [Google Scholar] [CrossRef] [PubMed]
- Tachibana, C. Transcriptomics today: Microarrays, RNA-seq, and more. Science 2015, 349, 544. [Google Scholar] [CrossRef]
- Zhao, S.; Fung-Leung, W.-P.; Bittner, A.; Ngo, K.; Liu, X. Comparison of RNA-Seq and Microarray in Transcriptome Profiling of Activated T Cells. PLoS ONE 2014, 9, e78644. [Google Scholar] [CrossRef]
- Liu, B.; Qian, S.B. Translational regulation in nutrigenomics. Adv. Nutr. 2011, 2, 511–519. [Google Scholar] [CrossRef] [PubMed]
- Tsuda, T.; Ueno, Y.; Yoshikawa, T.; Kojo, H.; Osawa, T. Microarray profiling of gene expression in human adipocytes in response to anthocyanins. Biochem. Pharmacol. 2006, 71, 1184–1197. [Google Scholar] [CrossRef] [PubMed]
- Van Erk, M.J.; Blom, W.A.M.; van Ommen, B.; Hendriks, H.F.J. High-protein and high-carbohydrate breakfasts differentially change the transcriptome of human blood cells. Am. J. Clin. Nutr. 2006, 84, 1233–1241. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qu, Z.; Li, W.; Fu, B. MicroRNAs in Autoimmune Diseases. BioMed Res. Int. 2014, 2014, 527895. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Evangelatos, G.; Fragoulis, G.E.; Koulouri, V.; Lambrou, G.I. MicroRNAs in rheumatoid arthritis: From pathogenesis to clinical impact. Autoimmun. Rev. 2019, 18, 102391. [Google Scholar] [CrossRef]
- Kura, B.; Parikh, M.; Slezak, J.; Pierce, G.N. The Influence of Diet on MicroRNAs that Impact Cardiovascular Disease. Molecules 2019, 24, 1509. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cui, J.; Zhou, B.; Ross, S.A.; Zempleni, J. Nutrition, microRNAs, and Human Health. Adv. Nutr. 2017, 8, 105–112. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, L.; Chen, T.; Yin, Y.; Zhang, C.-Y.; Zhang, Y.-L. Dietary microRNA—A Novel Functional Component of Food. Adv. Nutr. 2019, 10, 711–721. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chiang, K.; Shu, J.; Zempleni, J.; Cui, J. Dietary MicroRNA Database (DMD): An Archive Database and Analytic Tool for Food-Borne microRNAs. PLoS ONE 2015, 10, e0128089. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sarda, S.; Hannenhalli, S. Next-generation sequencing and epigenomics research: A hammer in search of nails. Genom. Inform. 2014, 12, 2–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Giacconi, R.; Malavolta, M.; Bürkle, A.; Moreno-Villanueva, M.; Franceschi, C.; Capri, M.; Slagboom, P.E.; Jansen, E.; Dollé, M.E.T.; Grune, T.; et al. Nutritional Factors Modulating Alu Methylation in an Italian Sample from The Mark-Age Study Including Offspring of Healthy Nonagenarians. Nutrients 2019, 11, 2986. [Google Scholar] [CrossRef] [Green Version]
- Brandt, B.; Rashidiani, S.; Bán, Á.; Rauch, T.A. DNA Methylation-Governed Gene Expression in Autoimmune Arthritis. Int. J. Mol. Sci. 2019, 20, 5646. [Google Scholar] [CrossRef] [Green Version]
- Carlos-Reyes, Á.; López-González, J.S.; Meneses-Flores, M.; Gallardo-Rincón, D.; Ruíz-García, E.; Marchat, L.A.; Astudillo-de la Vega, H.; Hernández de la Cruz, O.N.; López-Camarillo, C. Dietary Compounds as Epigenetic Modulating Agents in Cancer. Front. Genet. 2019, 10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gay, S.; Wilson, A.G. The emerging role of epigenetics in rheumatic diseases. Rheumatology 2014, 53, 406–414. [Google Scholar] [CrossRef] [Green Version]
- Kalea, A.Z.; Drosatos, K.; Buxton, J.L. Nutriepigenetics and cardiovascular disease. Curr. Opin. Clin. Nutr. Metab. Care 2018, 21, 252–259. [Google Scholar] [CrossRef]
- Burdge, G.C.; Hoile, S.P.; Lillycrop, K.A. Epigenetics: Are there implications for personalised nutrition? Curr. Opin. Clin. Nutr. Metab. Care 2012, 15, 442–447. [Google Scholar] [CrossRef]
- Van Dijk, E.L.; Jaszczyszyn, Y.; Naquin, D.; Thermes, C. The Third Revolution in Sequencing Technology. Trends Genet. TIG 2018, 34, 666–681. [Google Scholar] [CrossRef] [PubMed]
- Midha, M.K.; Wu, M.; Chiu, K.P. Long-read sequencing in deciphering human genetics to a greater depth. Hum. Genet. 2019, 138, 1201–1215. [Google Scholar] [CrossRef]
- Gong, L.; Wong, C.H.; Idol, J.; Ngan, C.Y.; Wei, C.L. Ultra-long Read Sequencing for Whole Genomic DNA Analysis. J. Vis. Exp. JOVE 2019. [Google Scholar] [CrossRef] [Green Version]
- Kraft, F.; Kurth, I. Long-read sequencing in human genetics. Med. Genet. 2019, 31, 198–204. [Google Scholar] [CrossRef] [Green Version]
- Oikonomopoulos, S.; Bayega, A.; Fahiminiya, S.; Djambazian, H.; Berube, P.; Ragoussis, J. Methodologies for Transcript Profiling Using Long-Read Technologies. Front. Genet. 2020, 11. [Google Scholar] [CrossRef] [PubMed]
- Quick, J. Ultra-Long Read Nanopore Sequencing Methods for Metagenomics. J. Biomol. Tech. 2019, 30, S63. [Google Scholar]
- Li, J.; Xu, H.; Sun, Z.; Hou, Q.; Kwok, L.-Y.; Laga, W.; Wang, Y.; Ma, H.; Yu, Z.; Menghe, B.; et al. Effect of dietary interventions on the intestinal microbiota of Mongolian hosts. Sci. Bull. 2016, 61, 1605–1614. [Google Scholar] [CrossRef] [Green Version]
- Singh, A. Tools for metabolomics. Nat. Methods 2020, 17, 24. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McGee, E.E.; Kiblawi, R.; Playdon, M.C.; Eliassen, A.H. Nutritional Metabolomics in Cancer Epidemiology: Current Trends, Challenges, and Future Directions. Curr. Nutr. Rep. 2019, 8, 187–201. [Google Scholar] [CrossRef]
- Johansson-Persson, A.; Barri, T.; Ulmius, M.; Onning, G.; Dragsted, L.O. LC-QTOF/MS metabolomic profiles in human plasma after a 5-week high dietary fiber intake. Anal. Bioanal. Chem. 2013, 405, 4799–4809. [Google Scholar] [CrossRef] [PubMed]
- McCombie, G.; Browning, L.M.; Titman, C.M.; Song, M.; Shockcor, J.; Jebb, S.A.; Griffin, J.L. Omega-3 oil intake during weight loss in obese women results in remodelling of plasma triglyceride and fatty acids. Metab. Off. J. Metab. Soc. 2009, 5, 363–374. [Google Scholar] [CrossRef] [Green Version]
- Van Velzen, E.J.; Westerhuis, J.A.; van Duynhoven, J.P.; van Dorsten, F.A.; Grün, C.H.; Jacobs, D.M.; Duchateau, G.S.; Vis, D.J.; Smilde, A.K. Phenotyping tea consumers by nutrikinetic analysis of polyphenolic end-metabolites. J. Proteome Res. 2009, 8, 3317–3330. [Google Scholar] [CrossRef] [PubMed]
- Redeuil, K.; Smarrito-Menozzi, C.; Guy, P.; Rezzi, S.; Dionisi, F.; Williamson, G.; Nagy, K.; Renouf, M. Identification of novel circulating coffee metabolites in human plasma by liquid chromatography-mass spectrometry. J. Chromatogr. A 2011, 1218, 4678–4688. [Google Scholar] [CrossRef]
- Collins, C.; McNamara, A.E.; Brennan, L. Role of metabolomics in identification of biomarkers related to food intake. Proc. Nutr. Soc. 2019, 78, 189–196. [Google Scholar] [CrossRef]
- Parker, S.J.; Raedschelders, K.; Van Eyk, J.E. Emerging proteomic technologies for elucidating context-dependent cellular signaling events: A big challenge of tiny proportions. Proteomics 2015, 15, 1486–1502. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Levy, E.; Slavov, N. Single cell protein analysis for systems biology. Essays Biochem. 2018, 62, 595–605. [Google Scholar] [CrossRef] [PubMed]
- Minakshi, P.; Kumar, R.; Ghosh, M.; Saini, H.M.; Ranjan, K.; Brar, B.; Prasad, G. Chapter 14—Single-Cell Proteomics: Technology and Applications. In Single-Cell Omics; Barh, D., Azevedo, V., Eds.; Academic Press: Cambridge, MA, USA, 2019; pp. 283–318. [Google Scholar] [CrossRef]
- Schweigert, F.J. Nutritional Proteomics: Methods and Concepts for Research in Nutritional Science. Ann. Nutr. Metab. 2007, 51, 99–107. [Google Scholar] [CrossRef]
- Ganesh, V.; Hettiarachchy, N.S. Nutriproteomics: A promising tool to link diet and diseases in nutritional research. Biochim. Biophys. Acta BBA Proteins Proteom. 2012, 1824, 1107–1117. [Google Scholar] [CrossRef]
- Vojdani, A.; Gushgari, L.R.; Vojdani, E. Interaction between food antigens and the immune system: Association with autoimmune disorders. Autoimmun. Rev. 2020, 19, 102459. [Google Scholar] [CrossRef] [PubMed]
- Tsuda, R.; Ozawa, T.; Kobayashi, E.; Hamana, H.; Taki, H.; Tobe, K.; Sugiyama, E.; Iwamoto, M.; Imura, J.; Kishi, H.; et al. Monoclonal antibody against citrullinated peptides obtained from rheumatoid arthritis patients reacts with numerous citrullinated microbial and food proteins. Arthritis Rheumatol. 2015, 67, 2020–2031. [Google Scholar] [CrossRef] [Green Version]
- Manasson, J.; Blank, R.B.; Scher, J.U. The microbiome in rheumatology: Where are we and where should we go? Ann. Rheum. Dis. 2020, 79, 727. [Google Scholar] [CrossRef] [Green Version]
- Segal, J.P.; Mullish, B.H.; Quraishi, M.N.; Acharjee, A.; Williams, H.R.T.; Iqbal, T.; Hart, A.L.; Marchesi, J.R. The application of omics techniques to understand the role of the gut microbiota in inflammatory bowel disease. Ther. Adv. Gastroenterol. 2019, 12. [Google Scholar] [CrossRef] [Green Version]
- De Filippis, F.; Pasolli, E.; Tett, A.; Tarallo, S.; Naccarati, A.; De Angelis, M.; Neviani, E.; Cocolin, L.; Gobbetti, M.; Segata, N.; et al. Distinct Genetic and Functional Traits of Human Intestinal Prevotella copri Strains Are Associated with Different Habitual Diets. Cell Host Microbe 2019, 25, 444–453.e3. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, G.; Zhang, C.; Wu, H.; Wang, R.; Shen, J.; Wang, L.; Zhao, Y.; Pang, X.; Zhang, X.; Zhao, L.; et al. Genomic Microdiversity of Bifidobacterium pseudocatenulatum Underlying Differential Strain-Level Responses to Dietary Carbohydrate Intervention. MBio 2017, 8, e02348-16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Malla, M.A.; Dubey, A.; Kumar, A.; Yadav, S.; Hashem, A.; Abd_Allah, E.F. Exploring the Human Microbiome: The Potential Future Role of Next-Generation Sequencing in Disease Diagnosis and Treatment. Front. Immunol. 2019, 9. [Google Scholar] [CrossRef] [PubMed]
- Tang, Z.-Z.; Chen, G.; Hong, Q.; Huang, S.; Smith, H.M.; Shah, R.D.; Scholz, M.; Ferguson, J.F. Multi-Omic Analysis of the Microbiome and Metabolome in Healthy Subjects Reveals Microbiome-Dependent Relationships Between Diet and Metabolites. Front. Genet. 2019, 10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tang, W.H.W.; Wang, Z.; Levison, B.S.; Koeth, R.A.; Britt, E.B.; Fu, X.; Wu, Y.; Hazen, S.L. Intestinal Microbial Metabolism of Phosphatidylcholine and Cardiovascular Risk. N. Engl. J. Med. 2013, 368, 1575–1584. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mills, S.; Stanton, C.; Lane, J.A.; Smith, G.J.; Ross, R.P. Precision Nutrition and the Microbiome, Part I: Current State of the Science. Nutrients 2019, 11, 923. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Slatko, B.E.; Gardner, A.F.; Ausubel, F.M. Overview of Next-Generation Sequencing Technologies. Curr. Protoc. Mol. Biol. 2018, 122, e59. [Google Scholar] [CrossRef] [PubMed]
- Hirst, M.; Marra, M.A. Next generation sequencing based approaches to epigenomics. Brief. Funct. Genom. 2010, 9, 455–465. [Google Scholar] [CrossRef]
- Verma, M.; Kumar, V. Chapter 11—Single-Cell Epigenomics: Technology and Applications. In Single-Cell Omics; Barh, D., Azevedo, V., Eds.; Academic Press: Cambridge, MA, USA, 2019; pp. 215–229. [Google Scholar] [CrossRef]
- Hussey, B.; Lindley, M.R.; Mastana, S.S. Omega 3 fatty acids, inflammation and DNA methylation: An overview. Clin. Lipidol. 2017, 12, 24–32. [Google Scholar] [CrossRef]
- Kocic, H.; Damiani, G.; Stamenkovic, B.; Tirant, M.; Jovic, A.; Tiodorovic, D.; Peris, K. Dietary compounds as potential modulators of microRNA expression in psoriasis. Adv. Chronic Dis. 2019, 10. [Google Scholar] [CrossRef] [Green Version]
- Tili, E.; Michaille, J.-J.; Adair, B.; Alder, H.; Limagne, E.; Taccioli, C.; Ferracin, M.; Delmas, D.; Latruffe, N.; Croce, C.M. Resveratrol decreases the levels of miR-155 by upregulating miR-663, a microRNA targeting JunB and JunD. Carcinogenesis 2010, 31, 1561–1566. [Google Scholar] [CrossRef] [PubMed]
- Marques-Rocha, J.L.; Milagro, F.I.; Mansego, M.L.; Zulet, M.A.; Bressan, J.; Martínez, J.A. Expression of inflammation-related miRNAs in white blood cells from subjects with metabolic syndrome after 8 wk of following a Mediterranean diet–based weight loss program. Nutrition 2016, 32, 48–55. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rao, V.S.; Srinivas, K.; Sujini, G.N.; Kumar, G.N.S. Protein-Protein Interaction Detection: Methods and Analysis. Int. J. Proteom. 2014, 2014, 147648. [Google Scholar] [CrossRef] [Green Version]
- Dürholz, K.; Hofmann, J.; Iljazovic, A.; Häger, J.; Lucas, S.; Sarter, K.; Strowig, T.; Bang, H.; Rech, J.; Schett, G.; et al. Dietary Short-Term Fiber Interventions in Arthritis Patients Increase Systemic SCFA Levels and Regulate Inflammation. Nutrients 2020, 12, 3207. [Google Scholar] [CrossRef]
- Murtaza, N.; Burke, L.M.; Vlahovich, N.; Charlesson, B.; O’Neill, H.M.; Ross, M.L.; Campbell, K.L.; Krause, L.; Morrison, M. Analysis of the Effects of Dietary Pattern on the Oral Microbiome of Elite Endurance Athletes. Nutrients 2019, 11, 614. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gibofsky, A. Epidemiology, pathophysiology, and diagnosis of rheumatoid arthritis: A Synopsis. Am. J. Manag. Care 2014, 20, S128–S135. [Google Scholar]
- Kurkó, J.; Besenyei, T.; Laki, J.; Glant, T.T.; Mikecz, K.; Szekanecz, Z. Genetics of rheumatoid arthritis—A comprehensive review. Clin. Rev. Allergy Immunol. 2013, 45, 170–179. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- MacGregor, A.J.; Snieder, H.; Rigby, A.S.; Koskenvuo, M.; Kaprio, J.; Aho, K.; Silman, A.J. Characterizing the quantitative genetic contribution to rheumatoid arthritis using data from twins. Arthritis Rheum. 2000, 43, 30–37. [Google Scholar] [CrossRef]
- Deane, K.D.; Demoruelle, M.K.; Kelmenson, L.B.; Kuhn, K.A.; Norris, J.M.; Holers, V.M. Genetic and environmental risk factors for rheumatoid arthritis. Best Pr. Res. Clin. Rheumatol. 2017, 31, 3–18. [Google Scholar] [CrossRef] [PubMed]
- Cutolo, M.; Nikiphorou, E. Don’t neglect nutrition in rheumatoid arthritis! RMD Open 2018, 4, e000591. [Google Scholar] [CrossRef] [Green Version]
- Kim, H.Y.; Kim, H.R.; Lee, S.H. Advances in systems biology approaches for autoimmune diseases. Immune Netw. 2014, 14, 73–80. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, F.; Wei, K.; Slowikowski, K.; Fonseka, C.Y.; Rao, D.A.; Kelly, S.; Goodman, S.M.; Tabechian, D.; Hughes, L.B.; Salomon-Escoto, K.; et al. Defining inflammatory cell states in rheumatoid arthritis joint synovial tissues by integrating single-cell transcriptomics and mass cytometry. Nat. Immunol. 2019, 20, 928–942. [Google Scholar] [CrossRef]
- Chen, Y.-J.; Chang, W.-A.; Wu, L.-Y.; Hsu, Y.-L.; Chen, C.-H.; Kuo, P.-L. Systematic Analysis of Differential Expression Profile in Rheumatoid Arthritis Chondrocytes Using Next-Generation Sequencing and Bioinformatics Approaches. Int. J. Med. Sci. 2018, 15, 1129–1142. [Google Scholar] [CrossRef] [Green Version]
- Platzer, A.; Nussbaumer, T.; Karonitsch, T.; Smolen, J.S.; Aletaha, D. Analysis of gene expression in rheumatoid arthritis and related conditions offers insights into sex-bias, gene biotypes and co-expression patterns. PLoS ONE 2019, 14, e0219698. [Google Scholar] [CrossRef] [Green Version]
- Stephenson, W.; Donlin, L.T.; Butler, A.; Rozo, C.; Bracken, B.; Rashidfarrokhi, A.; Goodman, S.M.; Ivashkiv, L.B.; Bykerk, V.P.; Orange, D.E.; et al. Single-cell RNA-seq of rheumatoid arthritis synovial tissue using low-cost microfluidic instrumentation. Nat. Commun. 2018, 9, 791. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, X.; Xia, S.; Fu, B. RNA-seq analysis of synovial fibroblasts in human rheumatoid arthritis. Mol. Med. Rep. 2014, 10, 241–247. [Google Scholar] [CrossRef]
- Suzuki, A.; Terao, C.; Yamamoto, K. Linking of genetic risk variants to disease-specific gene expression via multi-omics studies in rheumatoid arthritis. Semin. Arthritis Rheum. 2019, 49, S49–S53. [Google Scholar] [CrossRef] [PubMed]
- Giannopoulou, E.G.; Elemento, O.; Ivashkiv, L.B. Use of RNA sequencing to evaluate rheumatic disease patients. Arthritis Res. Ther. 2015, 17, 167. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goulielmos, G.N.; Zervou, M.I.; Myrthianou, E.; Burska, A.; Niewold, T.B.; Ponchel, F. Genetic data: The new challenge of personalized medicine, insights for rheumatoid arthritis patients. Gene 2016, 583, 90–101. [Google Scholar] [CrossRef] [PubMed]
- Soukup, T.; Hloch, K.; Doseděl, M.; Tebbens, J.D.; Nekvindová, J.; Šembera, Š.; Veleta, T.; Pávek, P.; Barvík, I. The influence of coffee intake and genetics on adenosine pathway in rheumatoid arthritis. Pharmacogenomics 2020, 21, 735–749. [Google Scholar] [CrossRef]
- Gan, R.W.; Young, K.A.; Zerbe, G.O.; Demoruelle, M.K.; Weisman, M.H.; Buckner, J.H.; Gregersen, P.K.; Mikuls, T.R.; O’Dell, J.R.; Keating, R.M.; et al. Lower omega-3 fatty acids are associated with the presence of anti-cyclic citrullinated peptide autoantibodies in a population at risk for future rheumatoid arthritis: A nested case-control study. Rheumatology 2016, 55, 367–376. [Google Scholar] [CrossRef] [Green Version]
- Tasaki, S.; Suzuki, K.; Kassai, Y.; Takeshita, M.; Murota, A.; Kondo, Y.; Ando, T.; Nakayama, Y.; Okuzono, Y.; Takiguchi, M.; et al. Multi-omics monitoring of drug response in rheumatoid arthritis in pursuit of molecular remission. Nat. Commun. 2018, 9, 2755. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Okada, Y.; Wu, D.; Trynka, G.; Raj, T.; Terao, C.; Ikari, K.; Kochi, Y.; Ohmura, K.; Suzuki, A.; Yoshida, S.; et al. Genetics of rheumatoid arthritis contributes to biology and drug discovery. Nature 2014, 506, 376–381. [Google Scholar] [CrossRef] [PubMed]
- Deane, K.D.; Norris, J.M.; Holers, V.M. Preclinical rheumatoid arthritis: Identification, evaluation, and future directions for investigation. Rheum. Dis. Clin. N. Am. 2010, 36, 213–241. [Google Scholar] [CrossRef] [Green Version]
- Lucchino, B.; Spinelli, F.R.; Iannuccelli, C.; Guzzo, M.P.; Conti, F.; Di Franco, M. Mucosa-Environment Interactions in the Pathogenesis of Rheumatoid Arthritis. Cells 2019, 8, 700. [Google Scholar] [CrossRef] [Green Version]
- Deane, K.D.; El-Gabalawy, H. Pathogenesis and prevention of rheumatic disease: Focus on preclinical RA and SLE. Nat. Rev. Rheumatol. 2014, 10, 212–228. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Korczowska, I. Rheumatoid arthritis susceptibility genes: An overview. World J. Orthop. 2014, 5, 544–549. [Google Scholar] [CrossRef] [PubMed]
- Saad, M.N.; Mabrouk, M.S.; Eldeib, A.M.; Shaker, O.G. Identification of rheumatoid arthritis biomarkers based on single nucleotide polymorphisms and haplotype blocks: A systematic review and meta-analysis. J. Adv. Res. 2016, 7, 1–16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Walsh, A.M.; Whitaker, J.W.; Huang, C.C.; Cherkas, Y.; Lamberth, S.L.; Brodmerkel, C.; Curran, M.E.; Dobrin, R. Integrative genomic deconvolution of rheumatoid arthritis GWAS loci into gene and cell type associations. Genome Biol. 2016, 17, 79. [Google Scholar] [CrossRef] [Green Version]
- Li, P.; Wang, X.; Guo, X.; Wen, Y.; Liu, L.; Liang, X.; Du, Y.; Wu, C.; Wang, S.; Zhang, F. Integrative analysis of genome-wide association study and expression quantitative trait loci datasets identified various immune cell-related pathways for rheumatoid arthritis. Ann. Hum. Genet. 2020, 84, 72–79. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Lin, X.; Yu, H. Identifying genes related with rheumatoid arthritis via system biology analysis. Gene 2015, 571, 97–106. [Google Scholar] [CrossRef]
- O’Shea, J.J.; Plenge, R. JAK and STAT signaling molecules in immunoregulation and immune-mediated disease. Immunity 2012, 36, 542–550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Diogo, D.; Okada, Y.; Plenge, R.M. Genome-wide association studies to advance our understanding of critical cell types and pathways in rheumatoid arthritis: Recent findings and challenges. Curr. Opin. Rheumatol. 2014, 26, 85–92. [Google Scholar] [CrossRef] [PubMed]
- Noack, M.; Miossec, P. Selected cytokine pathways in rheumatoid arthritis. Semin. Immunopathol. 2017, 39, 365–383. [Google Scholar] [CrossRef]
- Yap, H.-Y.; Tee, S.Z.-Y.; Wong, M.M.-T.; Chow, S.-K.; Peh, S.-C.; Teow, S.-Y. Pathogenic Role of Immune Cells in Rheumatoid Arthritis: Implications in Clinical Treatment and Biomarker Development. Cells 2018, 7, 161. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Crofford, L.J. The Role of COX-2 in Rheumatoid Arthritis Synovial Tissues. Arthritis Res. 2000, 1, S30. [Google Scholar] [CrossRef] [Green Version]
- Hansildaar, R.; Vedder, D.; Baniaamam, M.; Tausche, A.K.; Gerritsen, M.; Nurmohamed, M.T. Cardiovascular risk in inflammatory arthritis: Rheumatoid arthritis and gout. Lancet Rheumatol. 2020. [Google Scholar] [CrossRef]
- Cunnane, G.; Whitehead, A.S. Amyloid precursors and amyloidosis in rheumatoid arthritis. Best Pract. Res. Clin. Rheumatol. 1999, 13, 615–628. [Google Scholar] [CrossRef]
- DeMizio, D.J.; Geraldino-Pardilla, L.B. Autoimmunity and Inflammation Link to Cardiovascular Disease Risk in Rheumatoid Arthritis. Rheumatol. Ther. 2020, 7, 19–33. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Giles, J.T.; Danielides, S.; Szklo, M.; Post, W.S.; Blumenthal, R.S.; Petri, M.; Schreiner, P.J.; Budoff, M.; Detrano, R.; Bathon, J.M. Insulin resistance in rheumatoid arthritis: Disease-related indicators and associations with the presence and progression of subclinical atherosclerosis. Arthritis Rheumatol. 2015, 67, 626–636. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nakano, K.; Whitaker, J.W.; Boyle, D.L.; Wang, W.; Firestein, G.S. DNA methylome signature in rheumatoid arthritis. Ann. Rheum. Dis. 2013, 72, 110–117. [Google Scholar] [CrossRef] [PubMed]
- Karami, J.; Aslani, S.; Tahmasebi, M.N.; Mousavi, M.J.; Sharafat Vaziri, A.; Jamshidi, A.; Farhadi, E.; Mahmoudi, M. Epigenetics in rheumatoid arthritis; fibroblast-like synoviocytes as an emerging paradigm in the pathogenesis of the disease. Immunol. Cell Biol. 2020, 98, 171–186. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Glant, T.T.; Mikecz, K.; Rauch, T.A. Epigenetics in the pathogenesis of rheumatoid arthritis. BMC Med. 2014, 12, 35. [Google Scholar] [CrossRef] [Green Version]
- Mahendran, S.M.; Keystone, E.C.; Krawetz, R.J.; Liang, K.; Diamandis, E.P.; Chandran, V. Elucidating the endogenous synovial fluid proteome and peptidome of inflammatory arthritis using label-free mass spectrometry. Clin. Proteom. 2019, 16, 23. [Google Scholar] [CrossRef] [Green Version]
- Burska, A.; Boissinot, M.; Ponchel, F. Cytokines as biomarkers in rheumatoid arthritis. Mediat. Inflamm. 2014, 2014, 545493. [Google Scholar] [CrossRef]
- Aterido, A.; Tornero, J.; Blanco, F.; Fernández-Gutiérrez, B.; González, A.; Cañete, J.D.; Maymó, J.; Alperi-López, M.; Olivé, À.; Corominas, H.; et al. FRI0100 Multi-omics analysis identifies a gene signature associated with the clinical response to anti-tnf therapy in rheumatoid arthritis. J. Ann. Rheum. Dis. 2018, 77, 594. [Google Scholar] [CrossRef]
- Romão, V.C.; Vital, E.M.; Fonseca, J.E.; Buch, M.H. Right drug, right patient, right time: Aspiration or future promise for biologics in rheumatoid arthritis? Arthritis Res. Ther. 2017, 19, 239. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lindstrom, T.M.; Robinson, W.H. Biomarkers for rheumatoid arthritis: Making it personal. Scand. J. Clin. Lab. Investig. 2010, 242, 79–84. [Google Scholar] [CrossRef] [PubMed]
- Tao, W.; Concepcion, A.N.; Vianen, M.; Marijnissen, A.C.A.; Lafeber, F.P.G.J.; Radstake, T.R.D.J.; Pandit, A. Multi-omics and machine learning accurately predicts clinical response to Adalimumab and Etanercept therapy in patients with rheumatoid arthritis. Arthritis Rheumatol. 2021, 73, 212–222. [Google Scholar] [CrossRef]
- Singh, V.; Kalliolias, G.D.; Ostaszewski, M.; Veyssiere, M.; Pilalis, E.; Gawron, P.; Mazein, A.; Bonnet, E.; Petit-Teixeira, E.; Niarakis, A. RA-map: Building a state-of-the-art interactive knowledge base for rheumatoid arthritis. Database J. Biol. Databases Curation 2020, 2020. [Google Scholar] [CrossRef] [PubMed]
- Teodoro, A.J. Bioactive Compounds of Food: Their Role in the Prevention and Treatment of Diseases. Oxidative Med. Cell. Longev. 2019, 2019, 3765986. [Google Scholar] [CrossRef] [Green Version]
- Christensen, J.J.; Ulven, S.M.; Thoresen, M.; Westerman, K.; Holven, K.B.; Andersen, L.F. Associations between dietary patterns and gene expression pattern in peripheral blood mononuclear cells: A cross-sectional study. Nutr. Metab. Cardiovasc. Dis. 2020, 30, 2111–2122. [Google Scholar] [CrossRef]
- Nguyen, Y.; Salliot, C.; Gelot, A.; Gambaretti, J.; Mariette, X.; Boutron-Ruault, M.-C.; Seror, R. Mediterranean diet and risk of rheumatoid arthritis: Findings from the French E3N-EPIC cohort study. Arthritis Rheumatol. 2021, 73, 69–77. [Google Scholar] [CrossRef] [PubMed]
- Konstantinidou, V.; Covas, M.I.; Sola, R.; Fitó, M. Up-to date knowledge on the in vivo transcriptomic effect of the Mediterranean diet in humans. Mol. Nutr. Food Res. 2013, 57, 772–783. [Google Scholar] [CrossRef] [PubMed]
- Camargo, A.; Delgado-Lista, J.; Garcia-Rios, A.; Cruz-Teno, C.; Yubero-Serrano, E.M.; Perez-Martinez, P.; Gutierrez-Mariscal, F.M.; Lora-Aguilar, P.; Rodriguez-Cantalejo, F.; Fuentes-Jimenez, F.; et al. Expression of proinflammatory, proatherogenic genes is reduced by the Mediterranean diet in elderly people. Br. J. Nutr. 2012, 108, 500–508. [Google Scholar] [CrossRef] [PubMed]
- Yubero-Serrano, E.M.; Gonzalez-Guardia, L.; Rangel-Zuñiga, O.; Delgado-Lista, J.; Gutierrez-Mariscal, F.M.; Perez-Martinez, P.; Delgado-Casado, N.; Cruz-Teno, C.; Tinahones, F.J.; Villalba, J.M.; et al. Mediterranean diet supplemented with coenzyme Q10 modifies the expression of proinflammatory and endoplasmic reticulum stress-related genes in elderly men and women. J. Gerontol. Ser. A Biol. Sci. Med. Sci. 2012, 67, 3–10. [Google Scholar] [CrossRef] [Green Version]
- Arpón, A.; Riezu-Boj, J.I.; Milagro, F.I.; Marti, A.; Razquin, C.; Martínez-González, M.A.; Corella, D.; Estruch, R.; Casas, R.; Fitó, M.; et al. Adherence to Mediterranean diet is associated with methylation changes in inflammation-related genes in peripheral blood cells. J. Physiol. Biochem. 2016, 73, 445–455. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Davis, C.; Bryan, J.; Hodgson, J.; Murphy, K. Definition of the Mediterranean Diet; a Literature Review. Nutrients 2015, 7, 9139–9153. [Google Scholar] [CrossRef]
- Oliviero, F.; Spinella, P.; Fiocco, U.; Ramonda, R.; Sfriso, P.; Punzi, L. How the Mediterranean diet and some of its components modulate inflammatory pathways in arthritis. Swiss Med. Wkly. 2015, 145, w14190. [Google Scholar] [CrossRef] [PubMed]
- Barbosa, D.S.; Simão, A.C.; Dichi, I. Chapter 114—Use of Olive Oil in Patients with Rheumatoid Arthritis. In Olives and Olive Oil in Health and Disease Prevention; Preedy, V.R., Watson, R.R., Eds.; Academic Press: San Diego, CA, USA, 2010; pp. 1057–1064. [Google Scholar] [CrossRef]
- Aparicio-Soto, M.; Sánchéz-Hidalgo, M.; Cárdeno, A.; Lucena, J.M.; Gonzáléz-Escribano, F.; Castillo, M.J.; Alarcón-de-la-Lastra, C. The phenolic fraction of extra virgin olive oil modulates the activation and the inflammatory response of T cells from patients with systemic lupus erythematosus and healthy donors. Mol. Nutr. Food Res. 2017, 61. [Google Scholar] [CrossRef]
- Castañer, O.; Covas, M.I.; Khymenets, O.; Nyyssonen, K.; Konstantinidou, V.; Zunft, H.F.; de la Torre, R.; Muñoz-Aguayo, D.; Vila, J.; Fitó, M. Protection of LDL from oxidation by olive oil polyphenols is associated with a downregulation of CD40-ligand expression and its downstream products in vivo in humans. Am. J. Clin. Nutr. 2012, 95, 1238–1244. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, Y.; Walsh, A.M.; Fearon, U.; Smith, M.D.; Wechalekar, M.D.; Yin, X.; Cole, S.; Orr, C.; McGarry, T.; Canavan, M.; et al. CD40L-Dependent Pathway Is Active at Various Stages of Rheumatoid Arthritis Disease Progression. J. Immunol. 2017, 198, 4490. [Google Scholar] [CrossRef] [Green Version]
- Melguizo-Rodríguez, L.; Manzano-Moreno, F.J.; Illescas-Montes, R.; Ramos-Torrecillas, J.; Luna-Bertos, E.d.; Ruiz, C.; García-Martínez, O. Bone Protective Effect of Extra-Virgin Olive Oil Phenolic Compounds by Modulating Osteoblast Gene Expression. Nutrients 2019, 11, 1722. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kostoglou-Athanassiou, I.; Athanassiou, L.; Athanassiou, P. The Effect of Omega-3 Fatty Acids on Rheumatoid Arthritis. Mediterr. J. Rheumatol. 2020, 31, 190–194. [Google Scholar] [CrossRef] [PubMed]
- Lourdudoss, C.; Di Giuseppe, D.; Wolk, A.; Westerlind, H.; Klareskog, L.; Alfredsson, L.; van Vollenhoven, R.F.; Lampa, J. Dietary Intake of Polyunsaturated Fatty Acids and Pain in Spite of Inflammatory Control Among Methotrexate-Treated Early Rheumatoid Arthritis Patients. Arthritis Care Res. 2018, 70, 205–212. [Google Scholar] [CrossRef] [PubMed]
- Di Giuseppe, D.; Wallin, A.; Bottai, M.; Askling, J.; Wolk, A. Long-term intake of dietary long-chain n-3 polyunsaturated fatty acids and risk of rheumatoid arthritis: A prospective cohort study of women. Ann. Rheum. Dis. 2014, 73, 1949–1953. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shapiro, J.A.; Koepsell, T.D.; Voigt, L.F.; Dugowson, C.E.; Kestin, M.; Nelson, J.L. Diet and rheumatoid arthritis in women: A possible protective effect of fish consumption. Epidemiology 1996, 7, 256–263. [Google Scholar] [CrossRef]
- Gioxari, A.; Kaliora, A.C.; Marantidou, F.; Panagiotakos, D.P. Intake of ω-3 polyunsaturated fatty acids in patients with rheumatoid arthritis: A systematic review and meta-analysis. Nutrition 2018, 45, 114–124.e4. [Google Scholar] [CrossRef]
- Lee, A.L.; Park, Y. The association between n-3 polyunsaturated fatty acid levels in erythrocytes and the risk of rheumatoid arthritis in Korean women. Ann. Nutr. Metab. 2013, 63, 88–95. [Google Scholar] [CrossRef] [PubMed]
- Gan, R.W.; Demoruelle, M.K.; Deane, K.D.; Weisman, M.H.; Buckner, J.H.; Gregersen, P.K.; Mikuls, T.R.; O’Dell, J.R.; Keating, R.M.; Fingerlin, T.E.; et al. Omega-3 fatty acids are associated with a lower prevalence of autoantibodies in shared epitope-positive subjects at risk for rheumatoid arthritis. Ann. Rheum. Dis. 2017, 76, 147–152. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Basu, A.; Schell, J.; Scofield, R.H. Dietary fruits and arthritis. Food Funct. 2018, 9, 70–77. [Google Scholar] [CrossRef]
- Murakami, I.; Murakami, K.; Hashimoto, M.; Tanaka, M.; Ito, H.; Fujii, T.; Torii, M.; Ikeda, K.; Kuwabara, A.; Tanaka, K.; et al. Intake frequency of vegetables or seafoods negatively correlates with disease activity of rheumatoid arthritis. PLoS ONE 2020, 15, e0228852. [Google Scholar] [CrossRef] [PubMed]
- Alwarith, J.; Kahleova, H.; Rembert, E.; Yonas, W.; Dort, S.; Calcagno, M.; Burgess, N.; Crosby, L.; Barnard, N.D. Nutrition Interventions in Rheumatoid Arthritis: The Potential Use of Plant-Based Diets. A Review. Front. Nutr. 2019, 6, 141. [Google Scholar] [CrossRef]
- Comee, L.; Taylor, C.A.; Nahikian-Nelms, M.; Ganesan, L.P.; Krok-Schoen, J.L. Dietary patterns and nutrient intake of individuals with rheumatoid arthritis and osteoarthritis in the United States. Nutrition 2019, 67–68, 110533. [Google Scholar] [CrossRef] [Green Version]
- Bärebring, L.; Winkvist, A.; Gjertsson, I.; Lindqvist, H.M. Poor Dietary Quality Is Associated with Increased Inflammation in Swedish Patients with Rheumatoid Arthritis. Nutrients 2018, 10, 1535. [Google Scholar] [CrossRef] [Green Version]
- Hu, Y.; Sparks, J.A.; Malspeis, S.; Costenbader, K.H.; Hu, F.B.; Karlson, E.W.; Lu, B. Long-term dietary quality and risk of developing rheumatoid arthritis in women. Ann. Rheum. Dis. 2017, 76, 1357–1364. [Google Scholar] [CrossRef]
- Yang, C.M.; Chen, Y.W.; Chi, P.L.; Lin, C.C.; Hsiao, L.D. Resveratrol inhibits BK-induced COX-2 transcription by suppressing acetylation of AP-1 and NF-κB in human rheumatoid arthritis synovial fibroblasts. Biochem. Pharmacol. 2017, 132, 77–91. [Google Scholar] [CrossRef] [PubMed]
- Oh, Y.-C.; Kang, O.-H.; Choi, J.-G.; Chae, H.-S.; Lee, Y.-S.; Brice, O.-O.; Jung, H.J.; Hong, S.-H.; Lee, Y.-M.; Kwon, D.-Y. Anti-Inflammatory Effect of Resveratrol by Inhibition of IL-8 Production in LPS-Induced THP-1. Cells 2009, 37, 1203–1214. [Google Scholar] [CrossRef]
- Rajasekhar, M.; Olsson, A.M.; Steel, K.J.A.; Georgouli, M.; Ranasinghe, U.; Brender Read, C.; Frederiksen, K.S.; Taams, L.S. MicroRNA-155 contributes to enhanced resistance to apoptosis in monocytes from patients with rheumatoid arthritis. J. Autoimmun. 2017, 79, 53–62. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xiong, G.; Huang, Z.; Jiang, H.; Pan, Z.; Xie, J.; Wang, S. Inhibition of microRNA-21 decreases the invasiveness of fibroblast-like synoviocytes in rheumatoid arthritis via TGFβ/Smads signaling pathway. Iran. J. Basic Med. Sci. 2016, 19, 787–793. [Google Scholar] [PubMed]
- Li, X.; Tian, F.; Wang, F. Rheumatoid arthritis-associated microRNA-155 targets SOCS1 and upregulates TNF-α and IL-1β in PBMCs. Int. J. Mol. Sci. 2013, 14, 23910–23921. [Google Scholar] [CrossRef] [Green Version]
- Källberg, H.; Jacobsen, S.; Bengtsson, C.; Pedersen, M.; Padyukov, L.; Garred, P.; Frisch, M.; Karlson, E.W.; Klareskog, L.; Alfredsson, L. Alcohol consumption is associated with decreased risk of rheumatoid arthritis: Results from two Scandinavian case-control studies. Ann. Rheum. Dis. 2009, 68, 222–227. [Google Scholar] [CrossRef] [PubMed]
- Sundström, B.; Johansson, I.; Rantapää-Dahlqvist, S. Interaction between dietary sodium and smoking increases the risk for rheumatoid arthritis: Results from a nested case-control study. Rheumatology 2015, 54, 487–493. [Google Scholar] [CrossRef] [Green Version]
- Iris, M.; Tsou, P.-S.; Sawalha, A.H. Caffeine downregulates inflammatory pathways involved in autoimmunity. bioRxiv 2017, 241539. [Google Scholar] [CrossRef] [Green Version]
- Baghel, S.S.; Thakran, R.D.; Messi, C.; Kapoor, S.; Garg, S.; Kashyap, V.; Zaheer, Q.; Malaviya, A.N. SAT0746-HPR Coffee decreases methotrexate intolerance and increases its compliance in rheumatoid arthritis (RA): A study by rheumatology nurse counsellors. J. Ann. Rheum. Dis. 2018, 77, 1833–1834. [Google Scholar] [CrossRef]
- Van Bussel, I.P.G.; Jolink-Stoppelenburg, A.; De Groot, C.P.G.M.; Müller, M.R.; Afman, L.A. Differences in genome-wide gene expression response in peripheral blood mononuclear cells between young and old men upon caloric restriction. Genes Nutr. 2016, 11, 13. [Google Scholar] [CrossRef] [Green Version]
- Dao, M.C.; Sokolovska, N.; Brazeilles, R.; Affeldt, S.; Pelloux, V.; Prifti, E.; Chilloux, J.; Verger, E.O.; Kayser, B.D.; Aron-Wisnewsky, J.; et al. A Data Integration Multi-Omics Approach to Study Calorie Restriction-Induced Changes in Insulin Sensitivity. Front. Physiol. 2019, 9. [Google Scholar] [CrossRef]
- Roytblat, L.; Rachinsky, M.; Fisher, A.; Greemberg, L.; Shapira, Y.; Douvdevani, A.; Gelman, S. Raised interleukin-6 levels in obese patients. Obes. Res. 2000, 8, 673–675. [Google Scholar] [CrossRef] [PubMed]
- Hotamisligil, G.S.; Arner, P.; Caro, J.F.; Atkinson, R.L.; Spiegelman, B.M. Increased adipose tissue expression of tumor necrosis factor-alpha in human obesity and insulin resistance. J. Clin. Investig. 1995, 95, 2409–2415. [Google Scholar] [CrossRef] [PubMed]
- Gallagher, L.; Cregan, S.; Biniecka, M.; Cunningham, C.; Veale, D.J.; Kane, D.J.; Fearon, U.; Mullan, R.H. Insulin-Resistant Pathways Are Associated with Disease Activity in Rheumatoid Arthritis and Are Subject to Disease Modification Through Metabolic Reprogramming: A Potential Novel Therapeutic Approach. Arthritis Rheumatol. 2020, 72, 896–902. [Google Scholar] [CrossRef] [PubMed]
- Brenner, M.; Laragione, T.; Gulko, P.S. Short-term low-magnesium diet reduces autoimmune arthritis severity and synovial tissue gene expression. Physiol. Genom. 2017, 49, 238–242. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Zhai, L.; Wei, W. High-Methionine Diet Attenuates Severity of Arthritis and Modulates IGF-I Related Gene Expressions in an Adjuvant Arthritis Rats Model. Mediat. Inflamm. 2016, 2016, 9280529. [Google Scholar] [CrossRef] [PubMed]
- Imada, K.; Lin, N.; Liu, C.; Lu, A.; Chen, W.; Yano, M.; Sato, T.; Ito, A. Nobiletin, a citrus polymethoxy flavonoid, suppresses gene expression and production of aggrecanases-1 and -2 in collagen-induced arthritic mice. Biochem. Biophys. Res. Commun. 2008, 373, 181–185. [Google Scholar] [CrossRef] [PubMed]
- Norling, L.V.; Headland, S.E.; Dalli, J.; Arnardottir, H.H.; Haworth, O.; Jones, H.R.; Irimia, D.; Serhan, C.N.; Perretti, M. Proresolving and cartilage-protective actions of resolvin D1 in inflammatory arthritis. JCI Insight 2016, 1, e85922. [Google Scholar] [CrossRef] [PubMed]
- Lin, I.C.; Yamashita, S.; Murata, M.; Kumazoe, M.; Tachibana, H. Equol suppresses inflammatory response and bone erosion due to rheumatoid arthritis in mice. J. Nutr. Biochem. 2016, 32, 101–106. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, S. Green tea polyphenol epigallocatechin 3-gallate in arthritis: Progress and promise. Arthritis Res. 2010, 12, 208. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cassotta, M.; Pistollato, F.; Battino, M. Rheumatoid arthritis research in the 21st century: Limitations of traditional models, new technologies, and opportunities for a human biology-based approach. Altex 2020, 37, 223–242. [Google Scholar] [CrossRef] [PubMed]
- Steves, C.J.; Bird, S.; Williams, F.M.; Spector, T.D. The Microbiome and Musculoskeletal Conditions of Aging: A Review of Evidence for Impact and Potential Therapeutics. J. Bone Miner. Res. Off. J. Am. Soc. Bone Miner. Res. 2016, 31, 261–269. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Zhao, H.; Fan, D.; Liu, M.; Cao, J.; Xia, Y.; Ju, D.; Xiao, C.; Guan, Q. Interactions between Gut Microbiota and Immunomodulatory Cells in Rheumatoid Arthritis. Mediat. Inflamm. 2020, 2020, 1430605. [Google Scholar] [CrossRef]
- Van de Wiele, T.; Van Praet, J.T.; Marzorati, M.; Drennan, M.B.; Elewaut, D. How the microbiota shapes rheumatic diseases. Nat. Rev. Rheumatol. 2016, 12, 398–411. [Google Scholar] [CrossRef]
- Zhang, X.; Zhang, D.; Jia, H.; Feng, Q.; Wang, D.; Liang, D.; Wu, X.; Li, J.; Tang, L.; Li, Y.; et al. The oral and gut microbiomes are perturbed in rheumatoid arthritis and partly normalized after treatment. Nat. Med. 2015, 21, 895–905. [Google Scholar] [CrossRef] [PubMed]
- Zhong, D.; Wu, C.; Zeng, X.; Wang, Q. The role of gut microbiota in the pathogenesis of rheumatic diseases. Clin. Rheumatol. 2018, 37, 25–34. [Google Scholar] [CrossRef]
- Vaahtovuo, J.; Munukka, E.; Korkeamäki, M.; Luukkainen, R.; Toivanen, P. Fecal Microbiota in Early Rheumatoid Arthritis. J. Rheumatol. 2008, 35, 1500–1505. [Google Scholar]
- Chen, J.; Wright, K.; Davis, J.M.; Jeraldo, P.; Marietta, E.V.; Murray, J.; Nelson, H.; Matteson, E.L.; Taneja, V. An expansion of rare lineage intestinal microbes characterizes rheumatoid arthritis. Genome Med. 2016, 8, 43. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wells, P.M.; Adebayo, A.S.; Bowyer, R.C.E.; Freidin, M.B.; Finckh, A.; Strowig, T.; Lesker, T.R.; Alpizar-Rodriguez, D.; Gilbert, B.; Kirkham, B.; et al. Associations between gut microbiota and genetic risk for rheumatoid arthritis in the absence of disease. Lancet Rheumatol. 2020, 2, e418–e427. [Google Scholar] [CrossRef]
- Singh, R.K.; Chang, H.-W.; Yan, D.; Lee, K.M.; Ucmak, D.; Wong, K.; Abrouk, M.; Farahnik, B.; Nakamura, M.; Zhu, T.H.; et al. Influence of diet on the gut microbiome and implications for human health. J. Transl. Med. 2017, 15, 73. [Google Scholar] [CrossRef] [Green Version]
- Paolino, S.; Pacini, G.; Patanè, M.; Alessandri, E.; Cattelan, F.; Goegan, F.; Pizzorni, C.; Gotelli, E.; Cutolo, M. Interactions between microbiota, diet/nutrients and immune/inflammatory response in rheumatic diseases: Focus on rheumatoid arthritis. Reumatologia 2019, 57, 151–157. [Google Scholar] [CrossRef]
- Zhenyukh, O.; Civantos, E.; Ruiz-Ortega, M.; Sánchez, M.S.; Vázquez, C.; Peiró, C.; Egido, J.; Mas, S. High concentration of branched-chain amino acids promotes oxidative stress, inflammation and migration of human peripheral blood mononuclear cells via mTORC1 activation. Free Radic. Biol. Med. 2017, 104, 165–177. [Google Scholar] [CrossRef] [PubMed]
- Coras, R.; Murillo-Saich, J.D.; Guma, M. Circulating Pro- and Anti-Inflammatory Metabolites and Its Potential Role in Rheumatoid Arthritis Pathogenesis. Cells 2020, 9, 827. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wammers, M.; Schupp, A.-K.; Bode, J.G.; Ehlting, C.; Wolf, S.; Deenen, R.; Köhrer, K.; Häussinger, D.; Graf, D. Reprogramming of pro-inflammatory human macrophages to an anti-inflammatory phenotype by bile acids. Sci. Rep. 2018, 8, 255. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ratajczak, W.; Rył, A.; Mizerski, A.; Walczakiewicz, K.; Sipak, O.; Laszczyńska, M. Immunomodulatory potential of gut microbiome-derived short-chain fatty acids (SCFAs). Acta Biochim. Pol. 2019, 66, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Vinolo, M.A.R.; Rodrigues, H.G.; Nachbar, R.T.; Curi, R. Regulation of inflammation by short chain fatty acids. Nutrients 2011, 3, 858–876. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morrison, D.J.; Preston, T. Formation of short chain fatty acids by the gut microbiota and their impact on human metabolism. Gut Microbes 2016, 7, 189–200. [Google Scholar] [CrossRef] [Green Version]
- Li, M.; van Esch, B.C.A.M.; Henricks, P.A.J.; Folkerts, G.; Garssen, J. The Anti-inflammatory Effects of Short Chain Fatty Acids on Lipopolysaccharide- or Tumor Necrosis Factor α-Stimulated Endothelial Cells via Activation of GPR41/43 and Inhibition of HDACs. Front. Pharmacol. 2018, 9. [Google Scholar] [CrossRef] [Green Version]
- Chan, M.M.; Yang, X.; Wang, H.; Saaoud, F.; Sun, Y.; Fong, D. The Microbial Metabolite Trimethylamine N-Oxide Links Vascular Dysfunctions and the Autoimmune Disease Rheumatoid Arthritis. Nutrients 2019, 11, 1821. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pattison, D.J.; Symmons, D.P.; Lunt, M.; Welch, A.; Luben, R.; Bingham, S.A.; Khaw, K.T.; Day, N.E.; Silman, A.J. Dietary risk factors for the development of inflammatory polyarthritis: Evidence for a role of high level of red meat consumption. Arthritis Rheum. 2004, 50, 3804–3812. [Google Scholar] [CrossRef]
- Wang, Z.; Bergeron, N.; Levison, B.S.; Li, X.S.; Chiu, S.; Jia, X.; Koeth, R.A.; Li, L.; Wu, Y.; Tang, W.H.W.; et al. Impact of chronic dietary red meat, white meat, or non-meat protein on trimethylamine N-oxide metabolism and renal excretion in healthy men and women. Eur. Heart J. 2019, 40, 583–594. [Google Scholar] [CrossRef]
- Chen, B.; Zhao, Y.; Li, S.; Yang, L.; Wang, H.; Wang, T.; Bin, S.; Gai, Z.; Heng, X.; Zhang, C.; et al. Variations in oral microbiome profiles in rheumatoid arthritis and osteoarthritis with potential biomarkers for arthritis screening. Sci. Rep. 2018, 8, 17126. [Google Scholar] [CrossRef] [PubMed]
- Tong, Y.; Zheng, L.; Qing, P.; Zhao, H.; Li, Y.; Su, L.; Zhang, Q.; Zhao, Y.; Luo, Y.; Liu, Y. Oral Microbiota Perturbations Are Linked to High Risk for Rheumatoid Arthritis. Front. Cell. Infect. Microbiol. 2020, 9. [Google Scholar] [CrossRef] [PubMed]
- Lerner, A.; Aminov, R.; Matthias, T. Dysbiosis May Trigger Autoimmune Diseases via Inappropriate Post-Translational Modification of Host Proteins. Front. Microbial. 2016, 7. [Google Scholar] [CrossRef]
- Tomar, N.; Gupta, C.; Kaushik, M.; Wadhawan, A. Nutrigenomics: A perio-nutrition interrelationship. J. Oral Res. Rev. 2017, 9, 32–36. [Google Scholar]
- Singh, S.; Kalra, P. Nutrigenomics in periodontics—An overview. Int. J. Oral Health Dent. 2017, 3, 74–76. [Google Scholar] [CrossRef]
- Badsha, H. Role of Diet in Influencing Rheumatoid Arthritis Disease Activity. Open Rheumatol. J. 2018, 12, 19–28. [Google Scholar] [CrossRef] [Green Version]
- Forsyth, C.; Kouvari, M.; D’Cunha, N.M.; Georgousopoulou, E.N.; Panagiotakos, D.B.; Mellor, D.D.; Kellett, J.; Naumovski, N. The effects of the Mediterranean diet on rheumatoid arthritis prevention and treatment: A systematic review of human prospective studies. Rheumatol. Int. 2018, 38, 737–747. [Google Scholar] [CrossRef]
- Young, S.P.; Kapoor, S.R.; Viant, M.R.; Byrne, J.J.; Filer, A.; Buckley, C.D.; Kitas, G.D.; Raza, K. The impact of inflammation on metabolomic profiles in patients with arthritis. Arthritis Rheum. 2013, 65, 2015–2023. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, M.; Huang, J.; Fan, H.; He, D.; Zhao, S.; Shu, Y.; Li, H.; Liu, L.; Lu, S.; Xiao, C.; et al. Treatment of Rheumatoid Arthritis Using Combination of Methotrexate and Tripterygium Glycosides Tablets-A Quantitative Plasma Pharmacochemical and Pseudotargeted Metabolomic Approach. Front. Pharm. 2018, 9, 1051. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hedrick, V.E.; Dietrich, A.M.; Estabrooks, P.A.; Savla, J.; Serrano, E.; Davy, B.M. Dietary biomarkers: Advances, limitations and future directions. Nutr. J. 2012, 11, 109. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seok, J.; Warren, H.S.; Cuenca, A.G.; Mindrinos, M.N.; Baker, H.V.; Xu, W.; Richards, D.R.; McDonald-Smith, G.P.; Gao, H.; Hennessy, L.; et al. Genomic responses in mouse models poorly mimic human inflammatory diseases. Proc. Natl. Acad. Sci. USA 2013, 110, 3507–3512. [Google Scholar] [CrossRef] [Green Version]
- Fritsche, K.L. The science of fatty acids and inflammation. Adv. Nutr. 2015, 6, 293S–301S. [Google Scholar] [CrossRef] [PubMed]
- Cassotta, M.; Forbes-Hernández, T.Y.; Calderón Iglesias, R.; Ruiz, R.; Elexpuru Zabaleta, M.; Giampieri, F.; Battino, M. Links between Nutrition, Infectious Diseases, and Microbiota: Emerging Technologies and Opportunities for Human-Focused Research. Nutrients 2020, 12, 1827. [Google Scholar] [CrossRef] [PubMed]
- Karczewski, K.J.; Snyder, M.P. Integrative omics for health and disease. Nat. Rev. Genet. 2018, 19, 299–310. [Google Scholar] [CrossRef] [PubMed]
- Paul, B.J.; Kandy, H.I.; Krishnan, V. Pre-rheumatoid arthritis and its prevention. Eur. J. Rheumatol. 2017, 4, 161–165. [Google Scholar] [CrossRef]
- Pinu, F.R.; Beale, D.J.; Paten, A.M.; Kouremenos, K.; Swarup, S.; Schirra, H.J.; Wishart, D. Systems Biology and Multi-Omics Integration: Viewpoints from the Metabolomics Research Community. Metabolites 2019, 9, 76. [Google Scholar] [CrossRef] [Green Version]
- Fondi, M.; Liò, P. Multi -omics and metabolic modelling pipelines: Challenges and tools for systems microbiology. Microbiol. Res. 2015, 171, 52–64. [Google Scholar] [CrossRef] [PubMed]
- Dihazi, H.; Asif, A.R.; Beissbarth, T.; Bohrer, R.; Feussner, K.; Feussner, I.; Jahn, O.; Lenz, C.; Majcherczyk, A.; Schmidt, B.; et al. Integrative omics—From data to biology. Expert Rev. Proteom. 2018, 15, 463–466. [Google Scholar] [CrossRef] [PubMed]

| Approach | Subcategory | Targets | Techniques | References |
|---|---|---|---|---|
| Genomics | Genomics | Genes (DNA sequence) | Second generation: Illumina; SOLID; Ion Torrent. Third generation: PacBio; SMRT-seq; Illumina Tru-seq Synthetic Long-Read technology; Oxford Nanopore Technologies sequencing platform | [65,94] |
| Epigenomics | Modification of DNA and DNA- binding proteins | Whole-genome bisulfite sequencing (for DNA Methylation Analysis); ChIP-seq (for DNA–Protein Interaction Analysis); ATAC-Seq (for Chromatin Accessibility Analysis) | [95,96,97] | |
| Transcriptomics | Transcriptomics | mRNA | RNA-microarrays; RNA-seq: Illumina, SOLID, Ion Torrent (second generation); PacBio, SMRT-seq, Illumina TruSeq Synthetic Long-Read technology, Oxford Nanopore Technologies sequencing platform (third generation) | [47,48,49,50] |
| ncRNA-omics | non-coding RNA (including microRNA) | [57,98,99,100] | ||
| Proteomics | Proteomics | Proteins | Protein-microarrays; NMR Spectroscopy; MS; single-cell and ultrasensitive protein analyses | [79,80,81] |
| Interactomics | Protein-protein interaction, protein-small molecules interaction | TAP; Affinity Chromatography; Coimmunoprecipitation; Protein arrays; PFC; Phage display, NMR spectroscopy | [84,85,101] | |
| Metabolomics | Metabolomics | Metabolites | NMR; 1H NMR; MS; MALDI-TOF; SIMS; FTICR-MS | [72,73,74,75,76,77,78] |
| Lipidomics | Lipids | |||
| Aminomics | Aminoacids | |||
| Microbiomics | Microbiomics | Human Microbiota (including bacteria, fungi, protozoa, and viruses) | [87,88,89,90,91,102,103] | |
| Meta-genomics | Microbiota DNA | |||
| Meta-transcriptomics | Microbiota RNAs | |||
| Meta-proteomics | Microbiota proteins | |||
| Meta-bolomics | Microbiota metabolites |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cassotta, M.; Forbes-Hernandez, T.Y.; Cianciosi, D.; Elexpuru Zabaleta, M.; Sumalla Cano, S.; Dominguez, I.; Bullon, B.; Regolo, L.; Alvarez-Suarez, J.M.; Giampieri, F.; et al. Nutrition and Rheumatoid Arthritis in the ‘Omics’ Era. Nutrients 2021, 13, 763. https://doi.org/10.3390/nu13030763
Cassotta M, Forbes-Hernandez TY, Cianciosi D, Elexpuru Zabaleta M, Sumalla Cano S, Dominguez I, Bullon B, Regolo L, Alvarez-Suarez JM, Giampieri F, et al. Nutrition and Rheumatoid Arthritis in the ‘Omics’ Era. Nutrients. 2021; 13(3):763. https://doi.org/10.3390/nu13030763
Chicago/Turabian StyleCassotta, Manuela, Tamara Y. Forbes-Hernandez, Danila Cianciosi, Maria Elexpuru Zabaleta, Sandra Sumalla Cano, Irma Dominguez, Beatriz Bullon, Lucia Regolo, Josè Miguel Alvarez-Suarez, Francesca Giampieri, and et al. 2021. "Nutrition and Rheumatoid Arthritis in the ‘Omics’ Era" Nutrients 13, no. 3: 763. https://doi.org/10.3390/nu13030763
APA StyleCassotta, M., Forbes-Hernandez, T. Y., Cianciosi, D., Elexpuru Zabaleta, M., Sumalla Cano, S., Dominguez, I., Bullon, B., Regolo, L., Alvarez-Suarez, J. M., Giampieri, F., & Battino, M. (2021). Nutrition and Rheumatoid Arthritis in the ‘Omics’ Era. Nutrients, 13(3), 763. https://doi.org/10.3390/nu13030763