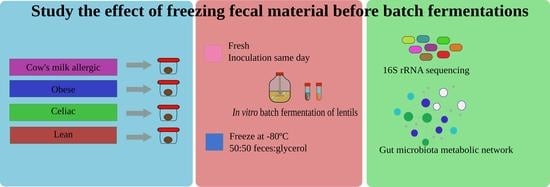
Effect of Freezing on Gut Microbiota Composition and Functionality for In Vitro Fermentation Experiments
Abstract
:1. Introduction
2. Materials and Methods
2.1. Samples
2.2. In Vitro Gastrointestinal Digestion of Lentils
2.3. Fecal Material Collection
2.4. In Vitro Fermentation
2.4.1. Stabilization
2.4.2. Lentils Batch Fermentation
2.5. 16S rRNA Gene Amplicon Sequencing and Bioinformatic Analysis
2.6. Functional Analysis Using Gut Microbiota Metabolic Networks
2.7. Statistical Analysis
3. Results
Effect of Freezing Fecal Samples on Gut Microbiota Community Structure
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Allin, K.H.; the IMI-DIRECT Consortium; Tremaroli, V.; Caesar, R.; Jensen, B.A.H.; Damgaard, M.T.F.; Bahl, M.I.; Licht, T.R.; Hansen, T.H.; Nielsen, T.; et al. Aberrant intestinal microbiota in individuals with prediabetes. Diabetologia 2018, 61, 810–820. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bultman, S.J. Interplay between diet, gut microbiota, epigenetic events, and colorectal cancer. Mol. Nutr. Food Res. 2017, 61, 1500902. [Google Scholar] [CrossRef] [Green Version]
- Ley, R.E.; Bäckhed, F.; Turnbaugh, P.; Lozupone, C.A.; Knight, R.D.; Gordon, J.I. Obesity alters gut microbial ecology. Proc. Natl. Acad. Sci. USA 2005, 102, 11070–11075. [Google Scholar] [CrossRef] [Green Version]
- Strati, F.; Cavalieri, D.; Albanese, D.; De Felice, C.; Donati, C.; Hayek, J.; Jousson, O.; Leoncini, S.; Renzi, D.; Calabrò, A.; et al. New evidences on the altered gut microbiota in autism spectrum disorders. Microbiome 2017, 5, 24. [Google Scholar] [CrossRef] [Green Version]
- Alam, M.T.; Amos, G.C.A.; Murphy, A.R.J.; Murch, S.; Wellington, E.M.H.; Arasaradnam, R.P. Microbial imbalance in inflammatory bowel disease patients at different taxonomic levels. Gut Pathog. 2020, 12, 1. [Google Scholar] [CrossRef] [PubMed]
- Bascuñán, K.A.; Araya, M.; Roncoroni, L.; Doneda, L.; Elli, L. Dietary Gluten as a Conditioning Factor of the Gut Microbiota in Celiac Disease. Adv. Nutr. 2019, 11, 160–174. [Google Scholar] [CrossRef]
- Bunyavanich, S.; Berin, M.C. Food allergy and the microbiome: Current understandings and future directions. J. Allergy Clin. Immunol. 2019, 144, 1468–1477. [Google Scholar] [CrossRef] [PubMed]
- Hughes, R.L.; Marco, M.L.; Hughes, J.P.; Keim, N.L.; Kable, M.E. The Role of the Gut Microbiome in Predicting Response to Diet and the Development of Precision Nutrition Models—Part I: Overview of Current Methods. Adv. Nutr. 2019, 10, 953–978. [Google Scholar] [CrossRef] [PubMed]
- Vandeputte, D.; Tito, R.Y.; VanLeeuwen, R.; Falony, G.; Raes, J. Practical considerations for large-scale gut microbiome studies. FEMS Microbiol. Rev. 2017, 41, S154–S167. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Hui, P.C.; Hui, M.; Yeoh, Y.K.; Wong, P.Y.; Chan, M.C.W.; Wong, M.C.S.; Ng, S.C.; Chan, F.K.L.; Chan, P.K.S. Impact of Preservation Method and 16S rRNA Hypervariable Region on Gut Microbiota Profiling. mSystems 2019, 4, e00271-18. [Google Scholar] [CrossRef] [Green Version]
- Cardona, S.; Eck, A.; Cassellas, M.; Gallart, M.; Alastrue, C.; Dore, J.; Azpiroz, F.; Roca, J.; Guarner, F.; Manichanh, C. Storage conditions of intestinal microbiota matter in metagenomic analysis. BMC Microbiol. 2012, 12, 158. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fouhy, F.; Deane, J.; Rea, M.; O’Sullivan, O.; Ross, R.P.; O’Callaghan, G.; Plant, B.J.; Stanton, C. The Effects of Freezing on Faecal Microbiota as Determined Using MiSeq Sequencing and Culture-Based Investigations. PLoS ONE 2015, 10, e0119355. [Google Scholar] [CrossRef] [PubMed]
- Cunningham, J.L.; Bramstång, L.; Singh, A.; Jayarathna, S.; Rasmusson, A.J.; Moazzami, A.; Müller, B. Impact of time and temperature on gut microbiota and SCFA composition in stool samples. PLoS ONE 2020, 15, e0236944. [Google Scholar] [CrossRef] [PubMed]
- Deschamps, C.; Fournier, E.; Uriot, O.; Lajoie, F.; Verdier, C.; Comtet-Marre, S.; Thomas, M.; Kapel, N.; Cherbuy, C.; Alric, M.; et al. Comparative methods for fecal sample storage to preserve gut microbial structure and function in an in vitro model of the human colon. Appl. Microbiol. Biotechnol. 2020, 104, 10233–10247. [Google Scholar] [CrossRef] [PubMed]
- Kia, E.; MacKenzie, B.W.; Middleton, D.; Lau, A.; Waite, D.W.; Lewis, G.; Chan, Y.-K.; Silvestre, M.; Cooper, G.; Poppitt, S.D.; et al. Integrity of the Human Faecal Microbiota following Long-Term Sample Storage. PLoS ONE 2016, 11, e0163666. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carroll, I.M.; Ringel-Kulka, T.; Siddle, J.P.; Klaenhammer, T.R.; Ringel, Y. Characterization of the Fecal Microbiota Using High-Throughput Sequencing Reveals a Stable Microbial Community during Storage. PLoS ONE 2012, 7, e46953. [Google Scholar] [CrossRef]
- Tedjo, D.I.; Jonkers, D.M.A.E.; Savelkoul, P.H.; Masclee, A.A.; van Best, N.; Pierik, M.J.; Penders, J. The Effect of Sampling and Storage on the Fecal Microbiota Composition in Healthy and Diseased Subjects. PLoS ONE 2015, 10, e0126685. [Google Scholar] [CrossRef]
- Han, M.; Hao, L.; Lin, Y.; Li, F.; Wang, J.; Yang, H.; Xiao, L.; Kristiansen, K.; Jia, H.; Li, J. A novel affordable reagent for room temperature storage and transport of fecal samples for metagenomic analyses. Microbiome 2018, 6, 43. [Google Scholar] [CrossRef]
- Ma, J.; Sheng, L.; Hong, Y.; Xi, C.; Gu, Y.; Zheng, N.; Li, M.; Chen, L.; Wu, G.; Li, Y.; et al. Variations of Gut Microbiome Profile Under Different Storage Conditions and Preservation Periods: A Multi-Dimensional Evaluation. Front. Microbiol. 2020, 11, 972. [Google Scholar] [CrossRef]
- Blekhman, R.; Tang, K.; Archie, E.A.; Barreiro, L.B.; Johnson, Z.P.; Wilson, M.E.; Kohn, J.; Yuan, M.L.; Gesquiere, L.; Grieneisen, L.E.; et al. Common methods for fecal sample storage in field studies yield consistent signatures of individual identity in microbiome sequencing data. Sci. Rep. 2016, 6, 31519. [Google Scholar] [CrossRef] [Green Version]
- Cammarota, G.; Ianiro, G.; Tilg, H.; Rajilic-Stojanovic, M.; Kump, P.; Satokari, R.; Sokol, H.; Arkkila, P.; Pintus, C.; Hart, A.; et al. European consensus conference on faecal microbiota transplantation in clinical practice. Gut 2017, 66, 569–580. [Google Scholar] [CrossRef] [PubMed]
- Brodkorb, A.; Egger, L.; Alminger, M.; Alvito, P.; Assunção, R.; Ballance, S.; Bohn, T.; Bourlieu-Lacanal, C.; Boutrou, R.; Carrière, F.; et al. INFOGEST static in vitro simulation of gastrointestinal food digestion. Nat. Protoc. 2019, 14, 991–1014. [Google Scholar] [CrossRef] [PubMed]
- Poeker, S.A.; Geirnaert, A.; Berchtold, L.; Greppi, A.; Krych, L.; Steinert, R.E.; De Wouters, T.; Lacroix, C. Understanding the prebiotic potential of different dietary fibers using an in vitro continuous adult fermentation model (PolyFermS). Sci. Rep. 2018, 8, 4318. [Google Scholar] [CrossRef]
- Perez-Burillo, S.; Rufián-Henares, J.; Pastoriza, S. Towards an improved global antioxidant response method (GAR+): Physiological-resembling in vitro digestion-fermentation method. Food Chem. 2018, 239, 1253–1262. [Google Scholar] [CrossRef] [PubMed]
- Klindworth, A.; Pruesse, E.; Schweer, T.; Peplies, J.; Quast, C.; Horn, M.; Glöckner, F.O. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 2013, 41, e1. [Google Scholar] [CrossRef] [PubMed]
- Callahan, B.J.; Mcmurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [Green Version]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [Green Version]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA Ribosomal RNA Gene Database Project: Improved Data Processing and Web-Based Tools. Nucleic Acids Res. 2012, 41, D590–D596. [Google Scholar] [CrossRef]
- Magnúsdóttir, S.; Heinken, A.; Kutt, L.; Ravcheev, D.; Bauer, E.; Noronha, A.; Greenhalgh, K.; Jäger, C.; Baginska, J.; Wilmes, P.; et al. Generation of genome-scale metabolic reconstructions for 773 members of the human gut microbiota. Nat. Biotechnol. 2017, 35, 81–89. [Google Scholar] [CrossRef]
- Gudmundsson, S.; Thiele, I. Computationally efficient flux variability analysis. BMC Bioinform. 2010, 11, 489. [Google Scholar] [CrossRef] [Green Version]
- Paliy, O.; Shankar, V. Application of multivariate statistical techniques in microbial ecology. Mol. Ecol. 2016, 25, 1032–1057. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deiana, P.; Santona, M.; Dettori, S.; Culeddu, N.; Dore, A.; Molinu, M.G. Multivariate approach to assess the chemical composition of Italian virgin olive oils as a function of variety and harvest period. Food Chem. 2019, 300, 125243. [Google Scholar] [CrossRef]
- Reichardt, N.; Vollmer, M.; Holtrop, G.; Farquharson, F.M.; Wefers, D.; Bunzel, M.; Duncan, S.H.; Drew, J.E.; Williams, L.M.; Milligan, G.; et al. Specific substrate-driven changes in human faecal microbiota composition contrast with functional redundancy in short-chain fatty acid production. ISME J. 2018, 12, 610–622. [Google Scholar] [CrossRef] [PubMed]
- Coyte, K.Z.; Schluter, J.; Foster, K.R. The ecology of the microbiome: Networks, competition, and stability. Science 2015, 350, 663–666. [Google Scholar] [CrossRef]
- Gomez, J.A.L.; Mukhopadhya, I.; Duncan, S.H.; Louis, P.; Shaw, S.; Collie-Duguid, E.; Crost, E.; Juge, N.; Flint, H.J. Formate cross-feeding and cooperative metabolic interactions revealed by transcriptomics in co-cultures of acetogenic and amylolytic human colonic bacteria. Environ. Microbiol. 2019, 21, 259–271. [Google Scholar] [CrossRef] [Green Version]
- Coyte, K.Z.; Rakoff-Nahoum, S. Understanding Competition and Cooperation within the Mammalian Gut Microbiome. Curr. Biol. 2019, 29, R538–R544. [Google Scholar] [CrossRef] [Green Version]
- Patnode, M.; Beller, Z.W.; Han, N.D.; Cheng, J.; Peters, S.L.; Terrapon, N.; Henrissat, B.; Le Gall, S.; Saulnier, L.; Hayashi, D.K.; et al. Interspecies Competition Impacts Targeted Manipulation of Human Gut Bacteria by Fiber-Derived Glycans. Cell 2019, 179, 59–73.e13. [Google Scholar] [CrossRef]
- Venema, K.; Abbeele, P.V.D. Experimental models of the gut microbiome. Best Pr. Res. Clin. Gastroenterol. 2013, 27, 115–126. [Google Scholar] [CrossRef]
- Wang, M.; Wichienchot, S.; He, X.; Fu, X.; Huang, Q.; Zhang, B. In vitro colonic fermentation of dietary fibers: Fermentation rate, short-chain fatty acid production and changes in microbiota. Trends Food Sci. Technol. 2019, 88, 1–9. [Google Scholar] [CrossRef]
- De Filippo, C.; Cavalieri, D.; Di Paola, M.; Ramazzotti, M.; Poullet, J.B.; Massart, S.; Collini, S.; Pieraccini, G.; Lionetti, P. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc. Natl. Acad. Sci. USA 2010, 107, 14691–14696. [Google Scholar] [CrossRef] [Green Version]
- Rowland, I.; Gibson, G.; Heinken, A.; Scott, K.; Swann, J.; Thiele, I.; Tuohy, K. Gut microbiota functions: Metabolism of nutrients and other food components. Eur. J. Nutr. 2018, 57, 1–24. [Google Scholar] [CrossRef] [Green Version]






Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pérez-Burillo, S.; Hinojosa-Nogueira, D.; Navajas-Porras, B.; Blasco, T.; Balzerani, F.; Lerma-Aguilera, A.; León, D.; Pastoriza, S.; Apaolaza, I.; Planes, F.J.; et al. Effect of Freezing on Gut Microbiota Composition and Functionality for In Vitro Fermentation Experiments. Nutrients 2021, 13, 2207. https://doi.org/10.3390/nu13072207
Pérez-Burillo S, Hinojosa-Nogueira D, Navajas-Porras B, Blasco T, Balzerani F, Lerma-Aguilera A, León D, Pastoriza S, Apaolaza I, Planes FJ, et al. Effect of Freezing on Gut Microbiota Composition and Functionality for In Vitro Fermentation Experiments. Nutrients. 2021; 13(7):2207. https://doi.org/10.3390/nu13072207
Chicago/Turabian StylePérez-Burillo, Sergio, Daniel Hinojosa-Nogueira, Beatriz Navajas-Porras, Telmo Blasco, Francesco Balzerani, Alberto Lerma-Aguilera, Daniel León, Silvia Pastoriza, Iñigo Apaolaza, Francisco J. Planes, and et al. 2021. "Effect of Freezing on Gut Microbiota Composition and Functionality for In Vitro Fermentation Experiments" Nutrients 13, no. 7: 2207. https://doi.org/10.3390/nu13072207
APA StylePérez-Burillo, S., Hinojosa-Nogueira, D., Navajas-Porras, B., Blasco, T., Balzerani, F., Lerma-Aguilera, A., León, D., Pastoriza, S., Apaolaza, I., Planes, F. J., Francino, M. P., & Rufián-Henares, J. Á. (2021). Effect of Freezing on Gut Microbiota Composition and Functionality for In Vitro Fermentation Experiments. Nutrients, 13(7), 2207. https://doi.org/10.3390/nu13072207