Gut Microbiota Differences According to Ultra-Processed Food Consumption in a Spanish Population
Abstract
:1. Introduction
2. Materials and Methods
2.1. Subjects
2.2. Assessment of Ultra-Processed Food Consumption
2.3. Fecal Sample Collection and Metagenomic Data
2.4. Anthropometric Measurements
2.5. Biochemical Measurements
2.6. Statistical Analysis
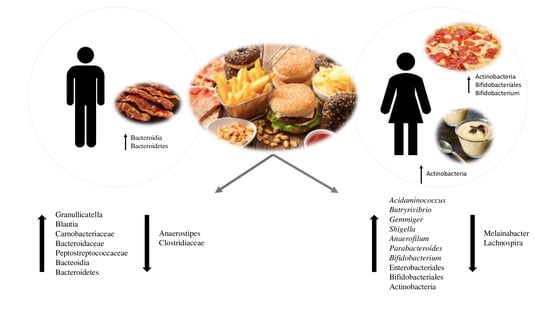
3. Results
3.1. Characteristics of the Population
3.2. Consumption of the Different Groups of Ultra-Processed Food
3.3. Analysis of Gut Microbiota Diversity According to Adjusted UPFs Consumption
3.4. Analysis of Gut Microbiota Composition According to UPF Consumption
3.5. Analysis of Associations between Bacterial Taxa and Groups of UPFs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hasan, N.; Yang, H. Factors affecting the composition of the gut microbiota, and its modulation. PeerJ 2019, 2019, 1–31. [Google Scholar] [CrossRef] [Green Version]
- Singh, R.K.; Chang, H.W.; Yan, D.; Lee, K.M.; Ucmak, D.; Wong, K.; Abrouk, M.; Farahnik, B.; Nakamura, M.; Zhu, T.H.; et al. Influence of diet on the gut microbiome and implications for human health. J. Transl. Med. 2017, 15, 1–17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koponen, K.K.; Salosensaari, A.; Ruuskanen, M.O.; Havulinna, A.S.; Männistö, S.; Jousilahti, P.; Palmu, J.; Salido, R.; Sanders, K.; Brennan, C.; et al. Associations of healthy food choices with gut microbiota profiles. Am. J. Clin. Nutr. 2021, 1–12. [Google Scholar] [CrossRef]
- Rauber, F.; Steele, E.M.; da Costa Louzada, M.L.; Millett, C.; Monteiro, C.A.; Levy, R.B. Ultra-processed food consumption and indicators of obesity in the United Kingdom population (2008–2016). PLoS ONE 2020, 15, e0232676. [Google Scholar] [CrossRef] [PubMed]
- Fardet, A.; Rock, E. Ultra-processed foods and food system sustainability: What are the links? Sustainability 2020, 12, 6280. [Google Scholar] [CrossRef]
- Monteiro, C.A.; Cannon, G.; Levy, R.B.; Moubarac, J.C.; Louzada, M.L.C.; Rauber, F.; Khandpur, N.; Cediel, G.; Neri, D.; Martinez-Steele, E.; et al. Ultra-processed foods: What they are and how to identify them. Public Health Nutr. 2019, 22, 936–941. [Google Scholar] [CrossRef]
- Marti, A. Ultra-processed foods are not “real food” but really affect your health. Nutrients 2019, 11, 1902. [Google Scholar] [CrossRef] [Green Version]
- De Deus Mendonça, R.; Souza Lopes, A.C.; Pimenta, A.M.; Gea, A.; Martinez-Gonzalez, M.A.; Bes-Rastrollo, M. Ultra-processed food consumption and the incidence of hypertension in a mediterranean cohort: The seguimiento universidad de navarra project. Am. J. Hypertens. 2017, 30, 358–366. [Google Scholar] [CrossRef] [Green Version]
- Bhurosy, T.; Kaschalk, E.; Smiley, A.; He, K. Comment on “ultraprocessed food consumption and risk of overweight and obesity: The University of Navarra Follow-Up (SUN) cohort study”. Am. J. Clin. Nutr. 2017, 105, 1012. [Google Scholar] [CrossRef]
- Martínez Steele, E.; Juul, F.; Neri, D.; Rauber, F.; Monteiro, C.A. Dietary share of ultra-processed foods and metabolic syndrome in the US adult population. Prev. Med. 2019, 125, 40–48. [Google Scholar] [CrossRef]
- Gómez-Donoso, C.; Sánchez-Villegas, A.; Martínez-González, M.A.; Gea, A.; Mendonça, R.d.D.; Lahortiga-Ramos, F.; Bes-Rastrollo, M. Ultra-processed food consumption and the incidence of depression in a Mediterranean cohort: The SUN Project. Eur. J. Nutr. 2020, 59, 1093–1103. [Google Scholar] [CrossRef]
- Srour, B.; Fezeu, L.K.; Kesse-Guyot, E.; Allès, B.; Debras, C.; Druesne-Pecollo, N.; Chazelas, E.; Deschasaux, M.; Hercberg, S.; Galan, P.; et al. Ultraprocessed Food Consumption and Risk of Type 2 Diabetes among Participants of the NutriNet-Santé Prospective Cohort. JAMA Intern. Med. 2020, 180, 283–291. [Google Scholar] [CrossRef] [PubMed]
- Senghor, B.; Sokhna, C.; Ruimy, R.; Lagier, J.C. Gut microbiota diversity according to dietary habits and geographical provenance. Hum. Microbiome J. 2018, 7–8, 1–9. [Google Scholar] [CrossRef]
- Cuevas-Sierra, A.; Riezu-Boj, J.I.; Guruceaga, E.; Milagro, F.I.; Martínez, J.A. Sex-Specific Associations between Gut Prevotellaceae and Host Genetics on Adiposity. Microorganisms 2020, 8, 938. [Google Scholar] [CrossRef]
- Monda, V.; Villano, I.; Messina, A.; Valenzano, A.; Esposito, T.; Moscatelli, F.; Viggiano, A.; Cibelli, G.; Chieffi, S.; Monda, M.; et al. Exercise modifies the gut microbiota with positive health effects. Oxid. Med. Cell. Longev. 2017, 2017, 3831972. [Google Scholar] [CrossRef] [PubMed]
- Mangiola, F.; Nicoletti, A.; Gasbarrini, A.; Ponziani, F.R. Gut microbiota and aging. Eur. Rev. Med. Pharmacol. Sci. 2018, 22, 7404–7413. [Google Scholar] [CrossRef]
- Santos-Marcos, J.A.; Haro, C.; Vega-Rojas, A.; Alcala-Diaz, J.F.; Molina-Abril, H.; Leon-Acuña, A.; Lopez-Moreno, J.; Landa, B.B.; Tena-Sempere, M.; Perez-Martinez, P.; et al. Sex differences in the gut microbiota as potential determinants of gender predisposition to disease. Mol. Nutr. Food Res. 2019, 1800870. [Google Scholar] [CrossRef]
- Yurkovetskiy, L.; Burrows, M.; Khan, A.A.; Graham, L.; Volchkov, P.; Becker, L.; Antonopoulos, D.; Umesaki, Y.; Chervonsky, A.V. Gender Bias in Autoimmunity is Influenced by Microbiota. Immunity 2013, 339, 1084–1088. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Markle, J.G.M.; Frank, D.N.; Mortin-Toth, S.; Robertson, C.E.; Feazel, L.M.; Rolle-Kampczyk, U.; Von Bergen, M.; McCoy, K.D.; Macpherson, A.J.; Danska, J.S. Sex differences in the gut microbiome drive hormone-dependent regulation of autoimmunity. Science 2013, 339, 1084–1088. [Google Scholar] [CrossRef] [Green Version]
- Org, E.; Mehrabian, M.; Parks, B.W.; Shipkova, P.; Liu, X.; Drake, T.A.; Lusis, A.J. Sex differences and hormonal effects on gut microbiota composition in mice. Gut Microbes. 2016, 7, 313–322. [Google Scholar] [CrossRef] [Green Version]
- Lay, C.; Rigottier-Gois, L.; Holmstrøm, K.; Rajilic, M.; Vaughan, E.E.; De Vos, W.M.; Collins, M.D.; Thiel, R.; Namsolleck, P.; Blaut, M.; et al. Colonic microbiota signatures across five northern European countries. Appl. Environ. Microbiol. 2005, 71, 4153–4155. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Filippo, C.; Cavalieri, D.; Di Paola, M.; Ramazzotti, M.; Poullet, J.B.; Massart, S.; Collini, S.; Pieraccini, G.; Lionetti, P. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc. Natl. Acad. Sci. USA 2010, 107, 14691–14696. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Asarian, L.; Geary, N. Sex differences in the physiology of eating. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2013, 305, R1215–R1267. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luzia Da Silva, C.; Sousa, A.G.; Pereira, L.; Leão Borges, S.; Macedo Da Costa, T.H. Usual consumption of ultra-processed foods and its association with sex, age, physical activity, and body mass index in adults living in Brasília City, Brazil. Rev. Bras. Epidemiol. 2021, 24, e210033. [Google Scholar] [CrossRef]
- Ramos-Lopez, O.; Riezu-Boj, J.I.; Milagro, F.I.; Goni, L.; Cuervo, M.; Martinez, J.A. Differential lipid metabolism outcomes associated with ADRB2 gene polymorphisms in response to two dietary interventions in overweight/obese subjects. Nutr. Metab. Cardiovasc. Dis. 2018, 28, 165–172. [Google Scholar] [CrossRef]
- Goni, L.; Riezu-Boj, J.I.; Milagro, F.I.; Corrales, F.J.; Ortiz, L.; Cuervo, M.; Martínez, J.A. Interaction between an ADCY3 genetic variant and two weight-lowering diets affecting body fatness and body composition outcomes depending on macronutrient distribution: A randomized trial. Nutrients 2018, 10, 789. [Google Scholar] [CrossRef]
- World Medical Association. World Medical Association Declaration of Helsinki: Ethical principles for medical research involving human subjects. J. Am. Coll. Dent. 2014, 81, 14–18. [Google Scholar] [CrossRef] [Green Version]
- Martin-Moreno, J.M.; Boyle, P.; Gorgojo, L.; Maisonneuve, P.; Fernandez-Rodriguez, J.C.; Salvini, S.; Willett, W.C. Development and Validation of a Food Frequency Questionnaire in Spain. Int. J. Epidemiol. 1993, 22, 512–519. [Google Scholar] [CrossRef]
- Moreiras, O.; Carbajal, A.; Cabrera, L.; Cuadrado, C. Tablas de Composicion de Alimentos (Ciencia y Tecnica); Piramide: Madrid, Spain, 2011. [Google Scholar]
- Cranston, J.M.; Crockett, A.J.; Moss, J.R.; Pegram, R.W.; Stocks, N.P. Ultra-Processed Food and Health Outcomes: A narrative review. Nutrients 2020, 12, 1955. [Google Scholar]
- Monteiro, C.A.; Cannon, G.; Levy, R.; Moubarac, J.-C.; Jaime, P.; Martins, A.P.; Canella, D.; Louzada, M.; Parra, D. NOVA. The star shines bright. World Nutr. 2016, 7, 28–38. [Google Scholar]
- Morea, N. Ultra-Processed Foods: Nova Classification | Food Compliance Solutions. Available online: https://regulatory.mxns.com/en/ultra-processed-foods-nova-classification (accessed on 25 June 2021).
- Ojeda-Rodríguez, A.; Zazpe, I.; Alonso-Pedrero, L.; Zalba, G.; Guillen-Grima, F.; Martinez-Gonzalez, M.A.; Marti, A. Association between diet quality indexes and the risk of short telomeres in an elderly population of the SUN project. Clin. Nutr. 2020, 39, 2487–2494. [Google Scholar] [CrossRef]
- Hildebrand, F.; Tadeo, R.; Voigt, A.Y.; Bork, P.; Raes, J. LotuS: An efficient and user-friendly OTU processing pipeline. Microbiome 2014, 2, 30. [Google Scholar] [CrossRef] [Green Version]
- Edgar, R.C. UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 2013, 10, 996–998. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. UCHIME2: Improved chimera prediction for amplicon sequencing. bioRxiv 2016. [Google Scholar] [CrossRef] [Green Version]
- Rideout, J.R.; He, Y.; Navas-Molina, J.A.; Walters, W.A.; Ursell, L.K.; Gibbons, S.M.; Chase, J.; McDonald, D.; Gonzalez, A.; Robbins-Pianka, A.; et al. Subsampled open-reference clustering creates consistent, comprehensive OTU definitions and scales to billions of sequences. PeerJ 2014, 2, e545. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ritari, J.; Salojärvi, J.; Lahti, L.; de Vos, W.M. Improved taxonomic assignment of human intestinal 16S rRNA sequences by a dedicated reference database. BMC Genom. 2015, 16, 1–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gentleman, R.C.; Carey, V.J.; Bates, D.M.; Bolstad, B.; Dettling, M.; Dudoit, S.; Ellis, B.; Gautier, L.; Ge, Y.; Gentry, J.; et al. Bioconductor: Open software development for computational biology and bioinformatics. Genome Biol. 2004, 5, 1–16. [Google Scholar] [CrossRef] [Green Version]
- World Health Organization. Body Mass Index. Available online: https://www.euro.who.int/en/health-topics/disease-prevention/nutrition/a-healthy-lifestyle/body-mass-indexbmi#:~:text=BMI%2C%20formerly%20called%20the%20Quetelet,have%20a%20BMI%20of%2022.9 (accessed on 25 June 2021).
- Whitworth, J.A.; Chalmers, J. World Health Organisation-International Society of Hypertension (WHO/ISH) hypertension guidelines. Clin. Exp. Hypertens. 2004, 26, 747–752. [Google Scholar] [CrossRef]
- Trichopoulou, A.; Costacou, T.; Bamia, C.; Trichopoulos, D. Adherence to a Mediterranean Diet and Survival in a Greek Population. N. Engl. J. Med. 2003, 348, 2599–2608. [Google Scholar] [CrossRef] [Green Version]
- Martínez-González, M.A.; López-Fontana, C.; Varo, J.J.; Sánchez-Villegas, A.; Martinez, J.A. Validation of the Spanish version of the physical activity questionnaire used in the Nurses’ Health Study and the Health Professionals’ Follow-up Study. Public Health Nutr. 2005, 8, 920–927. [Google Scholar] [CrossRef] [PubMed]
- Friedewald, W.T.; Levy, R.I.; Fredrickson, D.S. Estimation of the Concentration of Low-Density Lipoprotein Cholesterol in Plasma, Whithout Use of the Preparative Ultracentrifuge. Clin. Chem. 1972, 18, 499–502. [Google Scholar] [CrossRef] [PubMed]
- Matthews, D.R.; Hosker, J.P.; Rudenski, A.S.; Naylor, B.A.; Treacher, D.F.; Turner, R.C. Homeostasis model assessment: Insulin resistance and β-cell function from fasting plasma glucose and insulin concentrations in man. Diabetologia 1985, 28, 412–419. [Google Scholar] [CrossRef] [Green Version]
- Dhariwal, A.; Chong, J.; Habib, S.; King, I.L.; Agellon, L.B.; Xia, J. MicrobiomeAnalyst: A web-based tool for comprehensive statistical, visual and meta-analysis of microbiome data. Nucleic Acids Res. 2017, 45, W180–W188. [Google Scholar] [CrossRef]
- Pan American Health Organization of the World Health Organization. Ultra-Processed Food and Drink Products in Latin America: Trends, Impact on Obesity, Policy Implications; PAHO: Washington, DC, USA, 2015. [Google Scholar]
- Pagliai, G.; Dinu, M.; Madarena, M.P.; Bonaccio, M.; Iacoviello, L.; Sofi, F. Consumption of ultra-processed foods and health status: A systematic review and meta-analysis. Br. J. Nutr. 2020, 125, 308–318. [Google Scholar] [CrossRef]
- Rauber, F.; Chang, K.; Vamos, E.P.; da Costa Louzada, M.L.; Monteiro, C.A.; Millett, C.; Levy, R.B. Ultra-processed food consumption and risk of obesity: A prospective cohort study of UK Biobank. Eur. J. Nutr. 2021, 60, 2169–2180. [Google Scholar] [CrossRef] [PubMed]
- Adjibade, M.; Julia, C.; Allès, B.; Touvier, M.; Lemogne, C.; Srour, B.; Hercberg, S.; Galan, P.; Assmann, K.E.; Kesse-Guyot, E. Prospective association between ultra-processed food consumption and incident depressive symptoms in the French NutriNet-Santé cohort. BMC Med. 2019, 17, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Zheng, L.; Sun, J.; Yu, X.; Zhang, D. Ultra-Processed Food Is Positively Associated With Depressive Symptoms Among United States Adults. Front. Nutr. 2020, 7, 449. [Google Scholar] [CrossRef] [PubMed]
- Pestoni, G.; Habib, L.; Reber, E.; Rohrmann, S.; Staub, K.; Stanga, Z.; Faeh, D. Ultraprocessed Food Consumption is Strongly and Dose-Dependently Associated with Excess Body Weight in Swiss Women. Obesity 2021, 29, 601–609. [Google Scholar] [CrossRef]
- Nestares, T.; Martín-Masot, R.; Flor-Alemany, M.; Bonavita, A.; Maldonado, J.; Aparicio, V.A. Influence of Ultra-Processed Foods Consumption on Redox Status and Inflammatory Signaling in Young Celiac Patients. Nutrients 2021, 13, 156. [Google Scholar] [CrossRef]
- Hall, K.D.; Ayuketah, A.; Brychta, R.; Cai, H.; Cassimatis, T.; Chen, K.Y.; Chung, S.T. Ultra-Processed Diets Cause Excess Calorie Intake and Weight Gain: An Inpatient Randomized Controlled Trial of Ad Libitum Food Intake. Cell Metab. 2019, 30, 7–77. [Google Scholar] [CrossRef] [Green Version]
- Ivancovsky-Wajcman, D.; Fliss-Isakov, N.; Webb, M.; Bentov, I.; Shibolet, O.; Kariv, R.; Zelber-Sagi, S. Ultra-processed food is associated with features of metabolic syndrome and non-alcoholic fatty liver disease. Liver Int. 2021, liv.14996. [Google Scholar] [CrossRef]
- Srour, B.; Touvier, M. Ultra-processed foods and human health: What do we already know and what will further research tell us? EClinicalMedicine 2021, 32, 100747. [Google Scholar] [CrossRef]
- Agus, A.; Denizot, J.; Thévenot, J.; Martinez-Medina, M.; Massier, S.; Sauvanet, P.; Bernalier-Donadille, A.; Denis, S.; Hofman, P.; Bonnet, R.; et al. Western diet induces a shift in microbiota composition enhancing susceptibility to Adherent-Invasive E. coli infection and intestinal inflammation. Sci. Rep. 2016, 6, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Nogal, A.; Valdes, A.M.; Menni, C. The role of short-chain fatty acids in the interplay between gut microbiota and diet in cardio-metabolic health. Gut Microbes 2021, 13, 1–24. [Google Scholar] [CrossRef]
- Magne, F.; Gotteland, M.; Gauthier, L.; Zazueta, A.; Pesoa, S.; Navarrete, P.; Balamurugan, R. The firmicutes/bacteroidetes ratio: A relevant marker of gut dysbiosis in obese patients? Nutrients 2020, 12, 1474. [Google Scholar] [CrossRef] [PubMed]
- Noble, E.E.; Olson, C.A.; Davis, E.; Tsan, L.; Chen, Y.-W.; Schade, R.; Liu, C.; Suarez, A.; Jones, R.B.; Goran, M.I.; et al. The gut microbiome regulates memory function. bioRxiv 2020. [Google Scholar] [CrossRef]
- Baldelli, V.; Scaldaferri, F.; Putignani, L.; Del Chierico, F. The role of enterobacteriaceae in gut microbiota dysbiosis in inflammatory bowel diseases. Microorganisms 2021, 9, 697. [Google Scholar] [CrossRef] [PubMed]
- Di Rienzi, S.C.; Sharon, I.; Wrighton, K.C.; Koren, O.; Hug, L.A.; Thomas, B.C.; Goodrich, J.K.; Bell, J.T.; Spector, T.D.; Banfield, J.F.; et al. The human gut and groundwater harbor non-photosynthetic bacteria belonging to a new candidate phylum sibling to Cyanobacteria. eLife 2013, 2, 1102. [Google Scholar] [CrossRef] [PubMed]
- Shively, C.A.; Register, T.C.; Appt, S.E.; Clarkson, T.B.; Uberseder, B.; Clear, K.Y.J.; Wilson, A.S.; Chiba, A.; Tooze, J.A.; Cook, K.L. Consumption of Mediterranean versus Western Diet Leads to Distinct Mammary Gland Microbiome Populations HHS Public Access. Cell Rep. 2018, 25, 47–56. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dahl, W.J.; Rivero Mendoza, D.; Lambert, J.M. Diet, nutrients and the microbiome. Prog. Mol. Biol. Transl. Sci. 2020, 171, 237–263. [Google Scholar] [CrossRef]
- Roy, D. Technological aspects related to the use of bifidobacteria in dairy products. Lait 2005, 85, 39–56. [Google Scholar] [CrossRef] [Green Version]
- Vemuri, R.; Sylvia, K.E.; Klein, S.L.; Forster, S.C.; Plebanski, M.; Eri, R.; Flanagan, K.L. The microgenderome revealed: Sex differences in bidirectional interactions between the microbiota, hormones, immunity and disease susceptibility HHS Public Access. Semin. Immunopathol. 2019, 41, 265–275. [Google Scholar] [CrossRef]
- Ozato, N.; Saito, S.; Yamaguchi, T.; Katashima, M.; Tokuda, I.; Sawada, K.; Katsuragi, Y.; Kakuta, M.; Imoto, S.; Ihara, K.; et al. Blautia genus associated with visceral fat accumulation in adults 20–76 years of age. NPJ Biofilms Microbiomes 2019, 5, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Yang, Y.; Cai, Q.; Zheng, W.; Steinwandel, M.; Blot, W.J.; Shu, X.-O.; Long, J. Oral microbiome and obesity in a large study of low-income and African-American populations. J. Oral Microbiol. 2019, 11, 1650597. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lane, M.; Howland, G.; West, M.; Hockey, M.; Marx, W.; Loughman, A.; O’Hely, M.; Jacka, F.; Rocks, T. The effect of ultra-processed very low-energy diets on gut microbiota and metabolic outcomes in individuals with obesity: A systematic literature review. Obes. Res. Clin. Pract. 2020, 14, 197–204. [Google Scholar] [CrossRef] [PubMed]
- White, L.S.; Van den Bogaerde, J.; Kamm, M. The gut microbiota: Cause and cure of gut diseases. Med. J. Aust. 2018, 209, 312–317. [Google Scholar] [CrossRef]
- Chassaing, B.; Koren, O.; Goodrich, J.K.; Poole, A.C.; Srinivasan, S.; Ley, R.E.; Gewirtz, A.T. Dietary emulsifiers impact the mouse gut microbiota promoting colitis and metabolic syndrome. Naure 2015, 519, 92–96. [Google Scholar] [CrossRef] [Green Version]
- Baker, J.M.; Al-Nakkash, L.; Herbst-Kralovetz, M.M. Estrogen–gut microbiome axis: Physiological and clinical implications. Maturitas 2017, 103, 45–53. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Song, C.-H.; Kim, N.; Hee Nam, R.; In Choi, S.; Lee, H.-N.; Surh, Y.-J. 7β-estradiol supplementation changes gut microbiota diversity in intact and colorectal cancer-induced icR male mice. Sci. Rep. 2020, 10, 12283. [Google Scholar] [CrossRef]
- Martinez-Chacon, G.; Munukka, E.; Kumar, H.; Pietila, S.; Saarinen, N.; Toivonen, R.; Salminen, S.; Hanninen, A.; Strauss, L.; Makela, S. A link between sex hormones, obesity and gut microbiota. Endocr. Abstr. 2017, 49. [Google Scholar] [CrossRef]





| Whole Population | Women | Men | p Value (Women-Men < 3) 4 | p Value (Women-Men > 5) 5 | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Variables | <3 serv/d (n = 96) | >5 serv/d (n = 90) | p Value 1 | <3 serv/d (n = 57) | >5 serv/d (n = 66) | p Value 2 | <3 serv/d (n = 39) | >5 serv/d (n = 24) | p Value 3 | ||
| UPF consumption (serv/ d) | 2.0 ± 0.1 | 6.4 ± 0.2 | <0.001 | 2.0 ± 0.1 | 6.2 ± 0.1 | <0.001 | 2.0 ± 0.1 | 7.01 ± 0.52 | <0.001 | 0.76 | 0. 21 |
| Age (y) | 46 ± 1 | 43 ± 1 | 0.03 | 45 ± 1 | 43.5± 1 | 0.17 | 48 ± 1 | 43.5 ± 1.9 | 0.08 | 0.27 | 0.98 |
| Smoking | 20 | 24 | 0.54 | 11 | 15 | 0.68 | 9 | 9 | 0.98 | 0.47 | 0.23 |
| Alcohol habit | 61 | 51 | 0.36 | 30 | 30 | 0.93 | 30 | 21 | 0.74 | 0.98 | 0.16 |
| METs | 28.5 ± 1.5 | 22.5 ± 2.4 | 0.07 | 22.6 ± 2.5 | 18.1 ± 1.9 | 0.15 | 37.0 ± 3.8 | 34.5 ± 6.7 | 0.72 | <0.001 | 0.007 |
| Depression prevalence | 0 | 6 | 0.01 | 0 | 5 | 0.01 | 0 | 1 | 0.43 | 0.89 | 0.19 |
| Anxiety prevalence | 4 | 8 | 0.01 | 3 | 6 | 0.02 | 1 | 2 | 0.53 | 0.36 | 0.14 |
| Energy intake (kcal) | 2444 ± 50 | 3685 ± 97 | <0.001 | 2372 ± 61 | 3608 ± 107 | <0.001 | 2629 ± 83 | 3844 ± 195 | <0.001 | 0.02 | 0.25 |
| Energy from UPFs (%) | 10.1 ± 0.5 | 22.8 ± 1.1 | <0.001 | 8.7 ± 0.4 | 21.5 ± 1.3 | <0.001 | 12.1 ± 0.8 | 26.3 ± 2.0 | <0.001 | 0.002 | 0.06 |
| Adherence to MD | 8.0 ± 0.1 | 6.0 ± 0.1 | <0.001 | 7.0 ± 0.2 | 6.0 ± 0.2 | <0.001 | 7.0 ± 0.2 | 6.0 ± 0.3 | 0.006 | 0.77 | 0.78 |
| BMI baseline (kg/m2) | 29.2 ± 0.4 | 30.9 ± 0.4 | 0.02 | 29.5 ± 0.5 | 30.5 ± 0.6 | 0.19 | 29.5 ± 0.6 | 31.6 ± 0.6 | 0.02 | 0.99 | 0.25 |
| Weight (kg) | 81.1 ± 1.2 | 87.1 ± 1.5 | 0.002 | 77.5 ± 1.3 | 82.0 ± 1.7 | 0.04 | 90.6 ± 2.2 | 97.6 ± 2.2 | 0.03 | <0.001 | <0.001 |
| Waist circumference (cm) | 96 ± 1 | 101 ± 1 | 0.02 | 94 ± 1 | 97 ± 2 | 0.14 | 102 ± 2 | 108 ± 2 | 0.07 | 0.01 | 0.002 |
| Hip circumference (cm) | 108 ± 1 | 111 ± 1 | 0.01 | 108 ± 1 | 112 ± 1 | 0.03 | 106 ± 1 | 108 ± 1 | 0.13 | 0.11 | 0.05 |
| SBP (mmHg) | 126 ± 1 | 125 ± 2 | 0.71 | 122 ± 2 | 120 ± 2 | 0.33 | 135 ± 3 | 136 ± 3 | 0.81 | 0.001 | <0.001 |
| DBP (mmHg) | 78 ± 1 | 78 ± 1 | 0.86 | 76 ± 1 | 76 ± 1 | 0.91 | 84 ± 2 | 82 ± 2 | 0.44 | <0.001 | 0.002 |
| Fat mass (kg) | 28.6 ± 1.3 | 33.8 ± 1.4 | 0.01 | 29.2 ± 1.6 | 34.1 ± 1.9 | 0.05 | 27.2 ± 2.3 | 33.2 ± 2.0 | 0.06 | 0.51 | 0.75 |
| Visceral fat mass (kg) | 1.2 ± 0.08 | 1.3 ± 0.09 | 0.22 | 0.9 ± 0.06 | 1.0 ± 0.08 | 0.45 | 1.9 ± 0.2 | 2.1 ± 0.1 | 0.58 | <0.001 | <0.001 |
| Glucose (mg/dL) | 95 ± 1 | 94 ± 1 | 0.74 | 94 ± 2 | 92 ± 1 | 0.49 | 97 ± 2 | 98 ± 2 | 0.71 | 0.22 | 0.006 |
| Total cholesterol (mg/dL) | 211 ± 2 | 215 ± 4 | 0.44 | 211 ± 4 | 213 ± 5 | 0.77 | 210 ± 6 | 218 ± 6 | 0.34 | 0.89 | 0.51 |
| HDL-cholesterol (mg/dL) | 59 ± 1 | 55 ± 1 | 0.04 | 61 ± 1 | 59 ± 2 | 0.41 | 52 ± 1 | 46 ± 1 | 0.01 | <0.001 | <0.001 |
| LDL-cholesterol (mg/dL) | 64 ± 3 | 57 ± 4 | 0.22 | 63 ± 4 | 57 ± 5 | 0.37 | 67 ± 7 | 58 ± 6 | 0.35 | 0.59 | 0.92 |
| Triglycerides (mg/dL) | 86 ± 3 | 105 ± 6 | 0.004 | 84 ± 4 | 92 ± 5 | 0.23 | 92 ± 7 | 130 ± 12 | 0.006 | 0.29 | 0.001 |
| ALT (U/L) | 22 ± 1 | 23 ± 1 | 0.44 | 20 ± 1 | 19 ± 1 | 0.59 | 26 ± 2 | 31 ± 2 | 0.04 | 0.04 | <0.001 |
| AST (U/L) | 22 ± 1 | 21 ± 1 | 0.57 | 21 ± 1 | 20 ± 1 | 0.33 | 24 ± 1 | 24 ± 1 | 0.72 | 0.14 | <0.001 |
| Insulin (mU/L) | 7.3 ± 0.4 | 8.0 ± 0.4 | 0.36 | 7.5 ± 0.5 | 7.4 ± 0.5 | 0.93 | 7.0 ± 0.7 | 9.0 ± 1 | 0.09 | 0.59 | 0.13 |
| Adiponectin (µg/mL) | 12.3 ± 0.4 | 11.5 ± 0.4 | 0.21 | 13.7 ± 0.5 | 12.9 ± 0.5 | 0.35 | 8.9 ± 0.5 | 8.6 ± 0.5 | 0.67 | <0.001 | <0.001 |
| TNF (pg/mL) | 0.8 ± 0.02 | 0.9 ± 0.03 | 0.16 | 0.8 ± 0.03 | 0.8 ± 0.04 | 0.95 | 0.8 ± 0.02 | 1.0 ± 0.06 | 0.009 | 0.99 | 0.01 |
| Leptin (ng/mL) | 32.3 ± 2.2 | 35.1 ± 2.8 | 0.44 | 39.9 ± 2.6 | 45.3 ± 3.6 | 0.22 | 12.6 ± 1.6 | 14.1 ± 1.7 | 0.52 | <0.001 | <0.001 |
| HOMA-IR | 1.8 ± 0.1 | 1.8 ± 0.1 | 0.69 | 1.8 ± 0.2 | 1.7 ± 0.1 | 0.61 | 1.7 ± 0.2 | 2.2 ± 0.2 | 0.15 | 0.75 | 0.06 |
| CRP (µg/mL) | 2.4 ± 0.3 | 2.8 ± 0.3 | 0.31 | 2.6 ± 0.3 | 3.1 ± 0.4 | 0.41 | 1.8 ± 0.4 | 2.3 ± 0.3 | 0.41 | 0.21 | 0.24 |
| Women | Men | Women-Men < 3 | Women-Men > 5 | |||||
|---|---|---|---|---|---|---|---|---|
| Servings/day | <3 serv/d | >5 serv/d | p Value 1 | <3 serv/d | >5 serv/d | p Value 2 | p Value 3 | p Value 4 |
| Dairy consumption | 0.1 ± 0.01 | 0.3 ± 0.08 | <0.001 | 0.23 ± 0.04 | 0.16 ± 0.02 | <0.001 | 0.98 | 0.94 |
| Meat consumption | 0.7 ± 0.03 | 1.4 ± 0.1 | <0.001 | 0.7 ± 0.2 | 1.2 ± 0.1 | <0.001 | 0.61 | 0.02 |
| Cereals consumption | 0.02 ± 0.01 | 0.06 ± 0.02 | 0.28 | 0.13 ± 0.08 | 0.24± 0.12 | 0.21 | 0.69 | 0.34 |
| Pizza consumption | 0.07 ± 0.007 | 0.2 ± 0.05 | <0.001 | 0.1 ± 0.01 | 0.08 ± 0.02 | 0.64 | 0.31 | 0.09 |
| Margarine consumption | 0.05 ± 0.01 | 0.1 ± 0.04 | 0.48 | 0.16 ± 0.07 | 0.06 ± 0.03 | 0.005 | 0.07 | 0.58 |
| Fried consumption | 0.1 ± 0.009 | 0.2 ± 0.03 | <0.001 | 0.13 ± 0.03 | 0.24 ± 0.02 | 0.001 | 0.33 | 0.32 |
| Cookies consumption | 0.1 ± 0.02 | 0.7 ± 0.1 | <0.001 | 0.36 ± 0.15 | 0.76 ± 0.07 | <0.001 | 0.59 | 0.29 |
| Light products consumption | 0.07 ± 0.01 | 0.6 ± 0.1 | <0.001 | 0.07 ± 0.13 | 0.29 ± 0.03 | 0.06 | 0.96 | 0.66 |
| Ready-to-eat food consumption | 0.05 ± 0.006 | 0.08 ± 0.02 | 0.26 | 0.03 ± 0.009 | 0.08 ± 0.02 | 0.009 | 0.88 | 0.79 |
| Mayonnaise consumption | 0.05 ± 0.005 | 0.1 ± 0.02 | 0.04 | 0.08 ± 0.03 | 0.13 ± 0.02 | 0.02 | 0.61 | 0.11 |
| Alcohol consumption | 0.03 ± 0.008 | 0.03 ± 0.009 | 0.98 | 0.11 ± 0.03 | 0.22 ± 0.06 | 0.15 | 0.002 | <0.001 |
| Pastries consumption | 0.7 ± 0.05 | 2.4 ± 0.3 | <0.001 | 0.85 ± 0.15 | 2.2 ± 0.31 | <0.001 | 0.13 | 0.23 |
| SSB consumption | 0.09 ± 0.01 | 0.3 ± 0.07 | <0.001 | 0.19 ± 0.05 | 0.43 ± 0.13 | <0.001 | 0.16 | <0.001 |
| Bacteria Name | Log2FC | p Value | FDR |
|---|---|---|---|
| Genus | |||
| Gemmiger | 2.163 | 1.1 × 10−9 | 7.11 × 10−8 |
| Granulicatella | 1.759 | 6.4 × 10−7 | 1.98 × 10−5 |
| Parabacteroides | 0.969 | 1.9 × 10−4 | 0.002 |
| Shigella | 1.622 | 5.6 × 10−4 | 0.008 |
| Bifidobacterium | 1.075 | 7.0 × 10−4 | 0.008 |
| Anaerofilum | 0.786 | 0.001 | 0.01 |
| Lachsnopira | −1.034 | 0.003 | 0.02 |
| Roseburia | −0.746 | 0.003 | 0.02 |
| Cc_115 | 0.777 | 0.007 | 0.04 |
| Oxalobacter | 1.055 | 0.008 | 0.04 |
| Collinsella | 0.735 | 0.008 | 0.04 |
| Family | |||
| Carnobacteriacea | 1.772 | 4.69 × 10−7 | 1.54 × 10−5 |
| Oxalobacteraceae | 1.324 | 6.59 × 10−4 | 0.01 |
| Bifidobacteriaceae | 0.919 | 0.003 | 0.03 |
| Order | |||
| Bifidobacteriales | 1.125 | 3.81 × 10−4 | 0.006 |
| Pasteurellales | −1.180 | 0.005 | 0.04 |
| Class | |||
| Actinobacteria | 0.852 | 8.86 × 104 | 0.01 |
| Phylum | |||
| Actinobacteria | 0.852 | 8.86 × 10−4 | 0.01 |
| Bacterial Name | Log2FC | p Value | FDR |
|---|---|---|---|
| Genus | |||
| Acidaminococcus | 4.022 | 4.92 × 10−9 | 3.0 × 10−7 |
| Butyrivibrio | 2.899 | 4.17 × 10−7 | 1.3 × 10−5 |
| Gemmiger | 2.34 | 6.25 × 10−7 | 1.3 × 10−5 |
| Shigella | 2.171 | 2.14 × 10−4 | 0.003 |
| Anaerofilum | 1.228 | 3.4 × 10−4 | 0.004 |
| Parabacteroides | 1.018 | 0.002 | 0.02 |
| Melainabacter | −1.976 | 0.002 | 0.02 |
| Lachnospira | −1.321 | 0.003 | 0.02 |
| Bifidobacterium | 1.052 | 0.006 | 0.04 |
| Order | |||
| Enterobacteriales | 1.682 | 0.002 | 0.03 |
| Bifidobacteriales | 1.079 | 0.004 | 0.03 |
| Phylum | |||
| Actinobacteria | 0.860 | 0.006 | 0.04 |
| Bacterial Name | log2FC | p Value | FDR |
|---|---|---|---|
| Genus | |||
| Anaerostipes | −4.361 | 3.04 × 10−7 | 1.88 × 10−5 |
| Granullicatella | 3.019 | 7.94 ×10−6 | 2.46 × 10−4 |
| Blautia | 1.231 | 0.002 | 0.04 |
| Family | |||
| Carnobacteriaceae | 2.71 | 2.2 × 10−5 | 7.23 × 10−4 |
| Clostridiaceae | −1.313 | 0.002 | 0.03 |
| Bacteroidaceae | 1.023 | 0.002 | 0.03 |
| Peptostreptococcaceae | 1.443 | 0.005 | 0.04 |
| Class | |||
| Bacteroidia | 0.804 | 7.37 × 10−4 | 0.01 |
| Phylum | |||
| Bacteroidetes | 0.799 | 1.1 × 10−4 | 8.84 × 10−4 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cuevas-Sierra, A.; Milagro, F.I.; Aranaz, P.; Martínez, J.A.; Riezu-Boj, J.I. Gut Microbiota Differences According to Ultra-Processed Food Consumption in a Spanish Population. Nutrients 2021, 13, 2710. https://doi.org/10.3390/nu13082710
Cuevas-Sierra A, Milagro FI, Aranaz P, Martínez JA, Riezu-Boj JI. Gut Microbiota Differences According to Ultra-Processed Food Consumption in a Spanish Population. Nutrients. 2021; 13(8):2710. https://doi.org/10.3390/nu13082710
Chicago/Turabian StyleCuevas-Sierra, Amanda, Fermín I. Milagro, Paula Aranaz, Jose Alfredo Martínez, and José I. Riezu-Boj. 2021. "Gut Microbiota Differences According to Ultra-Processed Food Consumption in a Spanish Population" Nutrients 13, no. 8: 2710. https://doi.org/10.3390/nu13082710
APA StyleCuevas-Sierra, A., Milagro, F. I., Aranaz, P., Martínez, J. A., & Riezu-Boj, J. I. (2021). Gut Microbiota Differences According to Ultra-Processed Food Consumption in a Spanish Population. Nutrients, 13(8), 2710. https://doi.org/10.3390/nu13082710