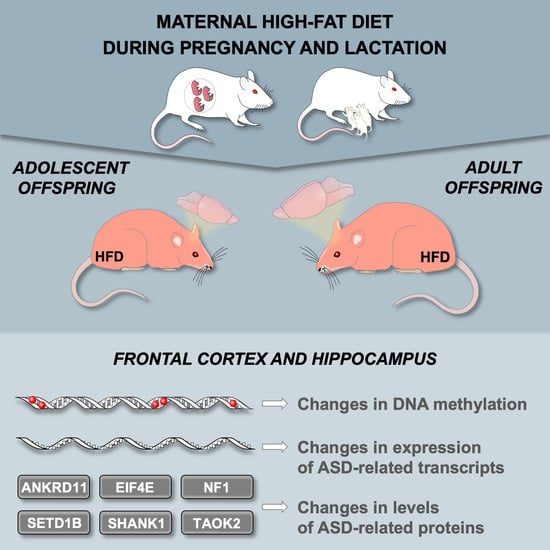
A Maternal High-Fat Diet during Early Development Provokes Molecular Changes Related to Autism Spectrum Disorder in the Rat Offspring Brain
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animals and Diets
2.2. Brain Tissue Collection
2.3. RNA Sequencing
2.4. Enzyme-Linked Immunosorbent Assay (ELISA)
2.5. Quantifying Global DNA Methylation
2.6. EpiTect Methyl II PCR Assay
2.7. Statistical Analysis
3. Results
3.1. Maternal HFD during Pregnancy and Lactation Changes the Expression of ASD-Related Genes in the Offspring Frontal Cortex
3.2. Maternal HFD during Pregnancy and Lactation Changes the Levels of ASD-Related Proteins in the Offspring Brain
3.3. Maternal HFD during Pregnancy and Lactation Changes Global DNA metHylation in the Offspring Brain
3.4. Maternal HFD during Pregnancy and Lactation Alters CpG Island DNA Methylation in the Offspring Brain
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kogan, M.D.; Vladutiu, C.J.; Schieve, L.A.; Ghandour, R.M.; Blumberg, S.J.; Zablotsky, B.; Perrin, J.M.; Shattuck, P.; Kuhlthau, K.A.; Harwood, R.L.; et al. The Prevalence of Parent-Reported Autism Spectrum Disorder among US Children. Pediatrics 2018, 142, e20174161. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jonsson, U.; Alaie, I.; Löfgren Wilteus, A.; Zander, E.; Marschik, P.B.; Coghill, D.; Bölte, S. Annual Research Review: Quality of life and childhood mental and behavioural disorders—A critical review of the research. J. Child Psychol. Psychiatry Allied Discip. 2017, 58, 439–469. [Google Scholar] [CrossRef]
- Eggebrecht, A.T.; Dworetsky, A.; Hawks, Z.; Coalson, R.; Adeyemo, B.; Davis, S.; Gray, D.; McMichael, A.; Petersen, S.E.; Constantino, J.N.; et al. Brain function distinguishes female carriers and non-carriers of familial risk for autism. Mol. Autism 2020, 11, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Rylaarsdam, L.; Guemez-Gamboa, A. Genetic Causes and Modifiers of Autism Spectrum Disorder. Front. Cell. Neurosci. 2019, 13, 385. [Google Scholar] [CrossRef]
- Le Belle, J.E.; Sperry, J.; Ngo, A.; Ghochani, Y.; Laks, D.R.; López-Aranda, M.; Silva, A.J.; Kornblum, H.I. Maternal inflammation contributes to brain overgrowth and autism-associated behaviors through altered redox signaling in stem and progenitor cells. Stem Cell Rep. 2014, 3, 725–734. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bölte, S.; Girdler, S.; Marschik, P.B. The contribution of environmental exposure to the etiology of autism spectrum disorder. Cell. Mol. Life Sci. 2019, 76, 1275–1297. [Google Scholar] [CrossRef] [Green Version]
- Sanchez, C.E.; Barry, C.; Sabhlok, A.; Russell, K.; Majors, A.; Kollins, S.H.; Fuemmeler, B.F. Maternal pre-pregnancy obesity and child neurodevelopmental outcomes: A meta-analysis. Obes. Rev. 2018, 19, 464–484. [Google Scholar] [CrossRef]
- Courchesne, E.; Pramparo, T.; Gazestani, V.H.; Lombardo, M.V.; Pierce, K.; Lewis, N.E. The ASD Living Biology: From cell proliferation to clinical phenotype. Mol. Psychiatry 2019, 24, 88–107. [Google Scholar] [CrossRef] [Green Version]
- Courchesne, E.; Gazestani, V.H.; Lewis, N.E. Prenatal Origins of ASD: The When, What, and How of ASD Development. Trends Neurosci. 2020, 43, 326–342. [Google Scholar] [CrossRef] [PubMed]
- Gawlińska, K.; Gawliński, D.; Filip, M.; Przegaliński, E. Relationship of maternal high-fat diet during pregnancy and lactation to offspring health. Nutr. Rev. 2021, 79, 709–725. [Google Scholar] [CrossRef] [PubMed]
- Keil, K.P.; Lein, P.J. DNA methylation: A mechanism linking environmental chemical exposures to risk of autism spectrum disorders? Environ. Epigenetics 2016, 2, dvv012. [Google Scholar] [CrossRef] [Green Version]
- Tremblay, M.W.; Jiang, Y.H. DNA methylation and susceptibility to autism spectrum disorder. Annu. Rev. Med. 2019, 70, 151–166. [Google Scholar] [CrossRef]
- McKee, S.E.; Zhang, S.; Chen, L.; Rabinowitz, J.D.; Reyes, T.M. Perinatal high fat diet and early life methyl donor supplementation alter one carbon metabolism and DNA methylation in the brain. J. Neurochem. 2018, 145, 362–373. [Google Scholar] [CrossRef] [PubMed]
- Indrio, F.; Martini, S.; Francavilla, R.; Corvaglia, L.; Cristofori, F.; Mastrolia, S.A.; Neu, J.; Rautava, S.; Spena, G.R.; Raimondi, F.; et al. Epigenetic matters: The link between early nutrition, microbiome, and long-term health development. Front. Pediatr. 2017, 5, 178. [Google Scholar] [CrossRef]
- Franzago, M.; Fraticelli, F.; Stuppia, L.; Vitacolonna, E. Nutrigenetics, epigenetics and gestational diabetes: Consequences in mother and child. Epigenetics 2019, 14, 215–235. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bansal, A.; Simmons, R.A. Epigenetics and developmental origins of diabetes: Correlation or causation? Am. J. Physiol.-Endocrinol. Metab. 2018, 315, E15–E28. [Google Scholar] [CrossRef] [Green Version]
- Li, Y. Epigenetic mechanisms link maternal diets and gut microbiome to obesity in the offspring. Front. Genet. 2018, 9, 342. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Xiao, X.; Zheng, J.; Li, M.; Yu, M.; Ping, F.; Wang, T.; Wang, X. A Maternal High-Fat Diet Induces DNA Methylation Changes That Contribute to Glucose Intolerance in Offspring. Front. Endocrinol. 2019, 10, 871. [Google Scholar] [CrossRef] [Green Version]
- Kong, L.; Chen, X.; Gissler, M.; Lavebratt, C. Relationship of prenatal maternal obesity and diabetes to offspring neurodevelopmental and psychiatric disorders: A narrative review. Int. J. Obes. 2020, 44, 1981–2000. [Google Scholar] [CrossRef] [PubMed]
- Gawliński, D.; Gawlińska, K.; Frankowska, M.; Filip, M. Maternal Diet Influences the Reinstatement of Cocaine-Seeking Behavior and the Expression of Melanocortin-4 Receptors in Female Offspring of Rats. Nutrients 2020, 12, 1462. [Google Scholar] [CrossRef]
- Paxinos, G.; Watson, C. The Rat Brain in Stereotaxic Coordinates, 4th ed.; Academic Press: San Diego, CA, USA, 1998. [Google Scholar]
- Gawlińska, K.; Gawliński, D.; Korostyński, M.; Borczyk, M.; Frankowska, M.; Piechota, M.; Filip, M.; Przegaliński, E. Maternal dietary patterns are associated with susceptibility to a depressive-like phenotype in rat offspring. Dev. Cogn. Neurosci. 2021, 47, 100879. [Google Scholar] [CrossRef]
- Abrahams, B.S.; Arking, D.E.; Campbell, D.B.; Mefford, H.C.; Morrow, E.M.; Weiss, L.A.; Menashe, I.; Wadkins, T.; Banerjee-Basu, S.; Packer, A. SFARI Gene 2.0: A community-driven knowledgebase for the autism spectrum disorders (ASDs). Mol. Autism 2013, 4, 1–3. [Google Scholar] [CrossRef] [Green Version]
- Chen, E.Y.; Tan, C.M.; Kou, Y.; Duan, Q.; Wang, Z.; Meirelles, G.V.; Clark, N.R.; Ma’ayan, A. Enrichr: Interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinform. 2013, 14, 1–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xie, Z.; Bailey, A.; Kuleshov, M.V.; Clarke, D.J.B.; Evangelista, J.E.; Jenkins, S.L.; Lachmann, A.; Wojciechowicz, M.L.; Kropiwnicki, E.; Jagodnik, K.M.; et al. Gene Set Knowledge Discovery with Enrichr. Curr. Protoc. 2021, 1, e90. [Google Scholar] [CrossRef] [PubMed]
- Gawlińska, K.; Gawliński, D.; Kowal-Wiśniewska, E.; Jarmuż-Szymczak, M.; Filip, M. Alteration of the Early Development Environment by Maternal Diet and the Occurrence of Autistic-like Phenotypes in Rat Offspring. Int. J. Mol. Sci. 2021, 22, 9662. [Google Scholar] [CrossRef]
- Quesnel-Vallières, M.; Weatheritt, R.J.; Cordes, S.P.; Blencowe, B.J. Autism spectrum disorder: Insights into convergent mechanisms from transcriptomics. Nat. Rev. Genet. 2019, 20, 51–63. [Google Scholar] [CrossRef]
- Cheatham, C.L. Nutritional Factors in Fetal and Infant Brain Development. Ann. Nutr. Metab. 2020, 75, 20–32. [Google Scholar] [CrossRef] [PubMed]
- Gawliński, D.; Gawlińska, K.; Frankowska, M.; Filip, M. Maternal high-sugar diet changes offspring vulnerability to reinstatement of cocaine-seeking behavior: Role of melanocortin-4 receptors. FASEB J. 2020, 34, 9192–9206. [Google Scholar] [CrossRef]
- Ergaz, Z.; Weinstein-Fudim, L.; Ornoy, A. Genetic and non-genetic animal models for autism spectrum disorders (ASD). Reprod. Toxicol. 2016, 64, 116–140. [Google Scholar] [CrossRef]
- Donovan, A.P.A.; Basson, M.A. The neuroanatomy of autism—A developmental perspective. J. Anat. 2017, 230, 4–15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hashem, S.; Nisar, S.; Bhat, A.A.; Yadav, S.K.; Azeem, M.W.; Bagga, P.; Fakhro, K.; Reddy, R.; Frenneaux, M.P.; Haris, M. Genetics of structural and functional brain changes in autism spectrum disorder. Transl. Psychiatry 2020, 10, 229. [Google Scholar] [CrossRef]
- Richards, R.; Greimel, E.; Kliemann, D.; Koerte, I.K.; Schulte-Körne, G.; Reuter, M.; Wachinger, C. Increased hippocampal shape asymmetry and volumetric ventricular asymmetry in autism spectrum disorder. NeuroImage Clin. 2020, 26, 102207. [Google Scholar] [CrossRef]
- Strange, B.A.; Witter, M.P.; Lein, E.S.; Moser, E.I. Functional organization of the hippocampal longitudinal axis. Nat. Rev. Neurosci. 2014, 15, 655–669. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Edlow, A.G.; Guedj, F.; Pennings, J.L.A.; Sverdlov, M.D.; Neri, C.; Bianchi, D.W. Males are from Mars, females are from Venus: Sex-specific fetal brain gene expression signatures in a mouse model of maternal diet-induced obesity. Am. J. Obstet. Gynecol. 2016, 214, 623.e1. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Robinson, E.B.; Lichtenstein, P.; Anckarsäter, H.; Happé, F.; Ronald, A. Examining and interpreting the female protective effect against autistic behavior. Proc. Natl. Acad. Sci. USA 2013, 110, 5258–5262. [Google Scholar] [CrossRef] [Green Version]
- Werling, D.M.; Geschwind, D.H. Sex differences in autism spectrum disorders. Curr. Opin. Neurol. 2013, 26, 146–153. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Li, N.; Li, C.; Zhang, Z.; Teng, H.; Wang, Y.; Zhao, T.; Shi, L.; Zhang, K.; Xia, K.; et al. Genetic evidence of gender difference in autism spectrum disorder supports the female-protective effect. Transl. Psychiatry 2020, 10, 4. [Google Scholar] [CrossRef] [Green Version]
- De Rubeis, S.; He, X.; Goldberg, A.P.; Poultney, C.S.; Samocha, K.; Cicek, A.E.; Kou, Y.; Liu, L.; Fromer, M.; Walker, S.; et al. Synaptic, transcriptional and chromatin genes disrupted in autism. Nature 2014, 515, 209–215. [Google Scholar] [CrossRef] [PubMed]
- Ayhan, F.; Konopka, G. Regulatory genes and pathways disrupted in autism spectrum disorders. Prog. Neuro-Psychopharmacol. Biol. Psychiatry 2019, 89, 57–64. [Google Scholar] [CrossRef]
- Sullivan, J.M.; De Rubeis, S.; Schaefer, A. Convergence of spectrums: Neuronal gene network states in autism spectrum disorder. Curr. Opin. Neurobiol. 2019, 59, 102–111. [Google Scholar] [CrossRef]
- Lugarà, E.; De Fusco, A.; Lignani, G.; Benfenati, F.; Humeau, Y. Synapsin I controls synaptic maturation of long-range projections in the lateral amygdala in a targeted selective fashion. Front. Cell. Neurosci. 2019, 13, 220. [Google Scholar] [CrossRef]
- Shih, P.Y.; Hsieh, B.Y.; Lin, M.H.; Huang, T.N.; Tsai, C.Y.; Pong, W.L.; Lee, S.P.; Hsueh, Y.P. CTTNBP2 Controls Synaptic Expression of Zinc-Related Autism-Associated Proteins and Regulates Synapse Formation and Autism-like Behaviors. Cell Rep. 2020, 31, 107700. [Google Scholar] [CrossRef] [PubMed]
- Fatemi, S.H.; Folsom, T.D.; Reutiman, T.J.; Thuras, P.D. Expression of GABAB Receptors Is Altered in Brains of Subjects with Autism. Cerebellum 2009, 8, 64–69. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Richter, M.; Murtaza, N.; Scharrenberg, R.; White, S.H.; Johanns, O.; Walker, S.; Yuen, R.K.C.; Schwanke, B.; Bedürftig, B.; Henis, M.; et al. Altered TAOK2 activity causes autism-related neurodevelopmental and cognitive abnormalities through RhoA signaling. Mol. Psychiatry 2019, 24, 1329–1350. [Google Scholar] [CrossRef] [Green Version]
- Shih, Y.T.; Huang, T.N.; Hu, H.T.; Yen, T.L.; Hsueh, Y.P. Vcp Overexpression and Leucine Supplementation Increase Protein Synthesis and Improve Fear Memory and Social Interaction of Nf1 Mutant Mice. Cell Rep. 2020, 31, 107835. [Google Scholar] [CrossRef]
- Shi, R.; Redman, P.; Ghose, D.; Hwang, H.; Liu, Y.; Ren, X.; Ding, L.J.; Liu, M.; Jones, K.J.; Xu, W. Shank Proteins Differentially Regulate Synaptic Transmission. Eneuro 2017, 4. [Google Scholar] [CrossRef] [Green Version]
- Collins, B.E.; Greer, C.B.; Coleman, B.C.; Sweatt, J.D. Histone H3 lysine K4 methylation and its role in learning and memory. Epigenetics Chromatin 2019, 12, 1–16. [Google Scholar] [CrossRef]
- Gallagher, D.; Voronova, A.; Zander, M.A.; Cancino, G.I.; Bramall, A.; Krause, M.P.; Abad, C.; Tekin, M.; Neilsen, P.M.; Callen, D.F.; et al. Ankrd11 is a chromatin regulator involved in autism that is essential for neural development. Dev. Cell 2015, 32, 31–42. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Medrano-Fernández, A.; Delgado-Garcia, J.M.; del Blanco, B.; Llinares, M.; Sánchez-Campusano, R.; Olivares, R.; Gruart, A.; Barco, A. The Epigenetic Factor CBP Is Required for the Differentiation and Function of Medial Ganglionic Eminence-Derived Interneurons. Mol. Neurobiol. 2019, 56, 4440–4454. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roston, A.; Evans, D.; Gill, H.; McKinnon, M.; Isidor, B.; Cogné, B.; Mwenifumbo, J.; Van Karnebeek, C.; An, J.; Jones, S.J.M.; et al. SETD1B -associated neurodevelopmental disorder. J. Med. Genet. 2021, 58, 196–204. [Google Scholar] [CrossRef]
- Scott, T.M.; Guo, H.; Eichler, E.E.; Rosenfeld, J.A.; Pang, K.; Liu, Z.; Lalani, S.; Bi, W.; Yang, Y.; Bacino, C.A.; et al. BAZ2B haploinsufficiency as a cause of developmental delay, intellectual disability, and autism spectrum disorder. Hum. Mutat. 2020, 41, 921–925. [Google Scholar] [CrossRef]
- Zhang, C.; Xu, L.; Zheng, X.; Liu, S.; Che, F. Role of Ash1l in Tourette syndrome and other neurodevelopmental disorders. Dev. Neurobiol. 2021, 81, 79–91. [Google Scholar] [CrossRef]
- Adegbola, A.; Musante, L.; Callewaert, B.; Maciel, P.; Hu, H.; Isidor, B.; Picker-Minh, S.; Le Caignec, C.; Chiaie, B.D.; Vanakker, O.; et al. Redefining the MED13L syndrome. Eur. J. Hum. Genet. 2015, 23, 1308–1317. [Google Scholar] [CrossRef] [Green Version]
- De Anda, F.C.; Rosario, A.L.; Durak, O.; Tran, T.; Gräff, J.; Meletis, K.; Rei, D.; Soda, T.; Madabhushi, R.; Ginty, D.D.; et al. Autism spectrum disorder susceptibility gene TAOK2 affects basal dendrite formation in the neocortex. Nat. Neurosci. 2012, 15, 1022–1031. [Google Scholar] [CrossRef]
- Klejman, M.P.; Zhao, X.; van Schaik, F.M.A.; Herr, W.; Timmers, H.T.M. Mutational analysis of BTAF1-TBP interaction: BTAF1 can rescue DNA-binding defective TBP mutants. Nucleic Acids Res. 2005, 33, 5426–5436. [Google Scholar] [CrossRef] [Green Version]
- Radio, F.C.; Pang, K.; Ciolfi, A.; Levy, M.A.; Hernández-García, A.; Pedace, L.; Pantaleoni, F.; Liu, Z.; de Boer, E.; Jackson, A.; et al. SPEN haploinsufficiency causes a neurodevelopmental disorder overlapping proximal 1p36 deletion syndrome with an episignature of X chromosomes in females. Am. J. Hum. Genet. 2021, 108, 502–516. [Google Scholar] [CrossRef] [PubMed]
- Yuen, R.K.C.; Merico, D.; Bookman, M.; Howe, J.L.; Thiruvahindrapuram, B.; Patel, R.V.; Whitney, J.; Deflaux, N.; Bingham, J.; Wang, Z.; et al. Whole genome sequencing resource identifies 18 new candidate genes for autism spectrum disorder. Nat. Neurosci. 2017, 20, 602–611. [Google Scholar] [CrossRef] [PubMed]
- Ansel, A.; Rosenzweig, J.P.; Zisman, P.D.; Melamed, M.; Gesundheit, B. Variation in gene expression in autism spectrum disorders: An extensive review of transcriptomic studies. Front. Neurosci. 2017, 10, 601. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chow, M.L.; Pramparo, T.; Winn, M.E.; Barnes, C.C.; Li, H.R.; Weiss, L.; Fan, J.B.; Murray, S.; April, C.; Belinson, H.; et al. Age-dependent brain gene expression and copy number anomalies in autism suggest distinct pathological processes at young versus mature ages. PLoS Genet. 2012, 8, e1002592. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ka, M.; Kim, W.Y. ANKRD11 associated with intellectual disability and autism regulates dendrite differentiation via the BDNF/TrkB signaling pathway. Neurobiol. Dis. 2018, 111, 138–152. [Google Scholar] [CrossRef]
- Hiraide, T.; Hattori, A.; Ieda, D.; Hori, I.; Saitoh, S.; Nakashima, M.; Saitsu, H. De novo variants in SETD1B cause intellectual disability, autism spectrum disorder, and epilepsy with myoclonic absences. Epilepsia Open 2019, 4, 476–481. [Google Scholar] [CrossRef] [Green Version]
- Fang, C.Y.; Lai, T.C.; Hsiao, M.; Chang, Y.C. The diverse roles of tao kinases in health and diseases. Int. J. Mol. Sci. 2020, 21, 7463. [Google Scholar] [CrossRef]
- Yadav, S.; Oses-Prieto, J.A.; Peters, C.J.; Zhou, J.; Pleasure, S.J.; Burlingame, A.L.; Jan, L.Y.; Jan, Y.N. TAOK2 Kinase Mediates PSD95 Stability and Dendritic Spine Maturation through Septin7 Phosphorylation. Neuron 2017, 93, 379–393. [Google Scholar] [CrossRef] [Green Version]
- Kahen, E.J.; Brohl, A.; Yu, D.; Welch, D.; Cubitt, C.L.; Lee, J.K.; Chen, Y.; Yoder, S.J.; Teer, J.K.; Zhang, Y.O.; et al. Neurofibromin level directs RAS pathway signaling and mediates sensitivity to targeted agents in malignant peripheral nerve sheath tumors. Oncotarget 2018, 9, 22571–22585. [Google Scholar] [CrossRef] [Green Version]
- Haebich, K.M.; Pride, N.A.; Walsh, K.S.; Chisholm, A.; Rouel, M.; Maier, A.; Anderson, V.; Barton, B.; Silk, T.; Korgaonkar, M.; et al. Understanding autism spectrum disorder and social functioning in children with neurofibromatosis type 1: Protocol for a cross-sectional multimodal study. BMJ Open 2019, 9, e030601. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weber, J.D.; Gutmann, D.H. Deconvoluting mTOR biology. Cell Cycle 2012, 11, 236–248. [Google Scholar] [CrossRef] [Green Version]
- Ganesan, H.; Balasubramanian, V.; Iyer, M.; Venugopal, A.; Subramaniam, M.D.; Cho, S.G.; Vellingiri, B. mTOR signalling pathway—A root cause for idiopathic autism? BMB Rep. 2019, 52, 424–433. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rosina, E.; Battan, B.; Siracusano, M.; Di Criscio, L.; Hollis, F.; Pacini, L.; Curatolo, P.; Bagni, C. Disruption of mTOR and MAPK pathways correlates with severity in idiopathic autism. Transl. Psychiatry 2019, 9, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.X.; Kim, G.H.; Tan, J.W.; Riso, A.E.; Sun, Y.; Xu, E.Y.; Liao, G.Y.; Xu, H.; Lee, S.H.; Do, N.Y.; et al. Elevated protein synthesis in microglia causes autism-like synaptic and behavioral aberrations. Nat. Commun. 2020, 11, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Monteiro, P.; Feng, G. SHANK proteins: Roles at the synapse and in autism spectrum disorder. Nat. Rev. Neurosci. 2017, 18, 147–157. [Google Scholar] [CrossRef]
- Sato, D.; Lionel, A.C.; Leblond, C.S.; Prasad, A.; Pinto, D.; Walker, S.; O’Connor, I.; Russell, C.; Drmic, I.E.; Hamdan, F.F.; et al. SHANK1 deletions in males with autism spectrum disorder. Am. J. Hum. Genet. 2012, 90, 879–887. [Google Scholar] [CrossRef] [Green Version]
- Hutsler, J.J.; Zhang, H. Increased dendritic spine densities on cortical projection neurons in autism spectrum disorders. Brain Res. 2010, 1309, 83–94. [Google Scholar] [CrossRef] [PubMed]
- Penzes, P.; Cahill, M.E.; Jones, K.A.; Vanleeuwen, J.E.; Woolfrey, K.M. Dendritic spine pathology in neuropsychiatric disorders. Nat. Neurosci. 2011, 14, 285–293. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gali Ramamoorthy, T.; Allen, T.J.; Davies, A.; Harno, E.; Sefton, C.; Murgatroyd, C.; White, A. Maternal overnutrition programs epigenetic changes in the regulatory regions of hypothalamic Pomc in the offspring of rats. Int. J. Obes. 2018, 42, 1431–1444. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wong, C.C.Y.; Smith, R.G.; Hannon, E.; Ramaswami, G.; Parikshak, N.N.; Assary, E.; Troakes, C.; Poschmann, J.; Schalkwyk, L.C.; Sun, W.; et al. Genome-wide DNA methylation profiling identifies convergent molecular signatures associated with idiopathic and syndromic autism in post-mortem human brain tissue. Hum. Mol. Genet. 2019, 28, 2201–2211. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chaturvedi, P.; Tyagi, S.C. Epigenetic mechanisms underlying cardiac degeneration and regeneration. Int. J. Cardiol. 2014, 173, 1–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]





| Frontal Cortex | Hippocampus | |||||||
|---|---|---|---|---|---|---|---|---|
| Males | Females | Males | Females | |||||
| PND 28 | PND 63 | PND 28 | PND 63 | PND 28 | PND 63 | PND 28 | PND 63 | |
| ANKRD11 | ↑ | − | − | − | − | − | − | − |
| EIF4E | ↑ | − | − | − | ↓ | ↓ | ↓ | − |
| NF1 | − | − | ↑ | − | − | − | − | − |
| SETD1B | ↑ | − | ↑ | − | − | − | − | − |
| SHANK1 | ↑ # | − | − | − | ↑ | − | − | − |
| TAOK2 | − | − | ↓ | − | − | ↓ | ↓ | − |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gawlińska, K.; Gawliński, D.; Borczyk, M.; Korostyński, M.; Przegaliński, E.; Filip, M. A Maternal High-Fat Diet during Early Development Provokes Molecular Changes Related to Autism Spectrum Disorder in the Rat Offspring Brain. Nutrients 2021, 13, 3212. https://doi.org/10.3390/nu13093212
Gawlińska K, Gawliński D, Borczyk M, Korostyński M, Przegaliński E, Filip M. A Maternal High-Fat Diet during Early Development Provokes Molecular Changes Related to Autism Spectrum Disorder in the Rat Offspring Brain. Nutrients. 2021; 13(9):3212. https://doi.org/10.3390/nu13093212
Chicago/Turabian StyleGawlińska, Kinga, Dawid Gawliński, Małgorzata Borczyk, Michał Korostyński, Edmund Przegaliński, and Małgorzata Filip. 2021. "A Maternal High-Fat Diet during Early Development Provokes Molecular Changes Related to Autism Spectrum Disorder in the Rat Offspring Brain" Nutrients 13, no. 9: 3212. https://doi.org/10.3390/nu13093212
APA StyleGawlińska, K., Gawliński, D., Borczyk, M., Korostyński, M., Przegaliński, E., & Filip, M. (2021). A Maternal High-Fat Diet during Early Development Provokes Molecular Changes Related to Autism Spectrum Disorder in the Rat Offspring Brain. Nutrients, 13(9), 3212. https://doi.org/10.3390/nu13093212