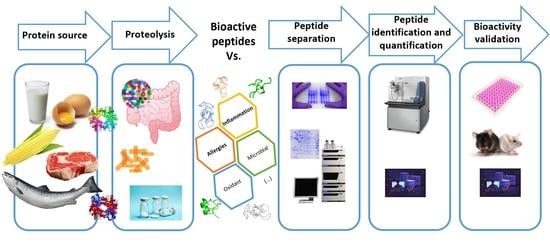
Proteomics Characterization of Food-Derived Bioactive Peptides with Anti-Allergic and Anti-Inflammatory Properties
Abstract
:1. Introduction
2. Bioactive Peptides with Anti-Allergic Properties
3. Bioactive Peptides with Anti-Inflammatory Properties
4. Bioactive Peptides by Proteomics
4.1. Recent Proteomic Approaches in the Identification and Quantification of Bioactive Peptides
4.2. In Silico Approaches for Bioactive Peptides
5. Concluding Remarks and Future Trends
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Korhonen, H.; Pihlanto, A. Bioactive peptides: Production and functionality. Int. Dairy J. 2006, 16, 945–960. [Google Scholar] [CrossRef]
- Daliri, E.B.M.; Oh, D.H.; Lee, B.H. Bioactive Peptides. Foods 2017, 6, 32. [Google Scholar] [CrossRef] [PubMed]
- Nicoletti, R.; Trincone, A. Bioactive Compounds Produced by Strains of Penicillium and Talaromyces of Marine Origin. Mar. Drugs 2016, 14, 37. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Srivastava, R.; Kulshreshtha, D.K. Bioactive polysaccharides from plants. Phytochemistry 1989, 28, 2877–2883. [Google Scholar] [CrossRef]
- Kalra, E.K. Nutraceutical-definition and introduction. AAPS PharmSci 2013, 5, 27–28. [Google Scholar] [CrossRef] [Green Version]
- Hasler, C.M. Regulation of Functional Foods and Nutraceuticals: A Global Perspective; IFT Press: Rome, Italy, 2005; ISBN 978-0-813-81177-2. [Google Scholar]
- Blaze, J. A Comparison of Current Regulatory Frameworks for Nutraceuticals in Australia, Canada, Japan, and the United States. Inov. Pharm. 2021, 12, 8. [Google Scholar] [CrossRef]
- Kitts, D.; Weiler, K. Bioactive Proteins and Peptides from Food Sources. Applications of Bioprocesses used in Isolation and Recovery. Curr. Pharm. Des. 2005, 9, 1309–1323. [Google Scholar] [CrossRef]
- Yates, A.A.; Erdman, J.W.; Shao, A.; Dolan, L.C.; Griffiths, J.C. Bioactive nutrients-Time for tolerable upper intake levels to address safety. Regul. Toxicol. Pharmacol. 2017, 84, 94–101. [Google Scholar] [CrossRef]
- Chakrabarti, S.; Guha, S.; Majumder, K. Food-Derived Bioactive Peptides in Human Health: Challenges and Opportunities. Nutrients 2018, 10, 1738. [Google Scholar] [CrossRef] [Green Version]
- Singh, B.P.; Vij, S.; Hati, S. Functional significance of bioactive peptides derived from soybean. Peptides 2014, 54, 171–179. [Google Scholar] [CrossRef]
- Sánchez, A.; Vázquez, A. Bioactive peptides: A review. Food Qual. Saf. 2017, 1, 29–46. [Google Scholar] [CrossRef]
- Manikkam, V.; Mathai, M.L.; Street, W.A.; Donkor, O.N.; Vasiljevic, T. Biofunctional and physicochemical properties of fish scales collagen-derived protein powders. Int. Food Res. J. 2016, 23, 1614–1622. [Google Scholar]
- Przybylski, R.; Bazinet, L.; Firdaous, L.; Kouach, M.; Goossens, J.F.; Dhulster, P.; Nedjar-Arroume, N. Electroseparation of Slaughterhouse By-Product: Antimicrobial Peptide Enrichment by pH Modification. Membranes 2020, 10, 90. [Google Scholar] [CrossRef]
- Lassoued, I.; Mora, L.; Barkia, A.; Aristoy, M.C.; Nasri, M.; Toldrá, F. Bioactive peptides identified in thornback ray skin’s gelatin hydrolysates by proteases from Bacillus subtilis and Bacillus amyloliquefaciens. J. Proteom. 2015, 128, 8–17. [Google Scholar] [CrossRef] [PubMed]
- Arrutia, F.; Puente, Á.; Riera, F.A.; Menéndez, C.; González, U.A. Influence of heat pre-treatment on BSA tryptic hydrolysis and peptide release. Food Chem. 2016, 202, 40–48. [Google Scholar] [CrossRef]
- Wang, W.; Gonzalez De Mejia, E. A New Frontier in Soy Bioactive Peptides that May Prevent Age-related Chronic Diseases. Compr. Rev. Food Sci. Food Saf. 2005, 4, 63–78. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.Y.; Je, J.Y.; Kim, S.K. Purification and characterization of antioxidant peptide from hoki (Johnius belengerii) frame protein by gastrointestinal digestion. J. Nutr. Biochem. 2007, 18, 31–38. [Google Scholar] [CrossRef]
- Ryan, J.T.; Ross, R.P.; Bolton, D.; Fitzgerald, G.F.; Stanton, C. Bioactive Peptides from Muscle Sources: Meat and Fish. Nutrients 2011, 3, 765–791. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ageitos, J.M.; Sánchez-Pérez, A.; Calo-Mata, P.; Villa, T.G. Antimicrobial peptides (AMPs): Ancient compounds that represent novel weapons in the fight against bacteria. Biochem. Pharmacol. 2017, 133, 117–138. [Google Scholar] [CrossRef]
- Chakrabarti, S.; Jahandideh, F.; Wu, J. Food-derived bioactive peptides on inflammation and oxidative stress. BioMed Res. Int. 2014, 2014. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guha, S.; Majumder, K. Structural-features of food-derived bioactive peptides with anti-inflammatory activity: A brief review. J. Food Biochem. 2019, 43, e12531. [Google Scholar] [CrossRef]
- Ahangari, H.; Yazdani, P.; Ebrahimi, V.; Soofiyani, S.R.; Azargun, R.; Tarhriz, V.; Eyvazi, S. An Updated review on production of food derived bioactive peptides; focus on the psychrotrophic bacterial proteases. Biocatal. Agric. Biotechnol. 2021, 35, 102051. [Google Scholar] [CrossRef]
- Chelliah, R.; Wei, S.; Daliri, E.B.M.; Elahi, F.; Yeon, S.J.; Tyagi, A.; Liu, S.; Madar, I.H.; Sultan, G.; Oh, D.H. The Role of Bioactive Peptides in Diabetes and Obesity. Foods 2021, 10, 2220. [Google Scholar] [CrossRef]
- Wu, S.; Bekhit, A.E.D.A.; Wu, Q.; Chen, M.; Liao, X.; Wang, J.; Ding, Y. Bioactive peptides and gut microbiota: Candidates for a novel strategy for reduction and control of neurodegenerative diseases. Trends Food Sci. Technol. 2021, 108, 164–176. [Google Scholar] [CrossRef]
- Aloo, S.O.; Oh, D.H. The Functional Interplay between Gut Microbiota, Protein Hydrolysates/Bioactive Peptides, and Obesity: A Critical Review on the Study Advances. Antioxidants 2022, 11, 333. [Google Scholar] [CrossRef]
- Zeng, M.; Cui, W.; Zhao, Y.; Liu, Z.; Dong, S.; Guo, Y. Antiviral active peptide from oyster. Chinese J. Oceanol. Limnol. 2008, 26, 307–312. [Google Scholar] [CrossRef]
- Gill, H.S.; Doull, F.; Rutherfurd, K.J.; Cross, M.L. Immunoregulatory peptides in bovine milk. Br. J. Nutr. 2000, 84, 111–117. [Google Scholar] [CrossRef] [PubMed]
- Sauer, S.; Luge, T. Nutriproteomics: Facts, concepts, and perspectives. Proteomics 2015, 15, 997–1013. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schrader, M.; Schulz-Knappe, P.; Fricker, L.D. Historical perspective of peptidomics. EuPA Open Proteomics 2014, 3, 171–182. [Google Scholar] [CrossRef] [Green Version]
- Capriotti, A.L.; Cavaliere, C.; Piovesana, S.; Samperi, R.; Laganà, A. Recent trends in the analysis of bioactive peptides in milk and dairy products. Anal. Bioanal. Chem. 2016, 408, 2677–2685. [Google Scholar] [CrossRef] [PubMed]
- Guerrero, A.; Dallas, D.C.; Contreras, S.; Chee, S.; Parker, E.A.; Sun, X.; Dimapasoc, L.; Barile, D.; German, J.B.; Lebrilla, C.B. Mechanistic peptidomics: Factors that dictate specificity in the formation of endogenous peptides in human milk. Mol. Cell. Proteom. 2014, 13, 3343–3351. [Google Scholar] [CrossRef]
- Ulug, S.K.; Jahandideh, F.; Wu, J. Novel technologies for the production of bioactive peptides. Trends Food Sci. Technol. 2021, 108, 27–39. [Google Scholar] [CrossRef]
- Renz, H.; Allen, K.J.; Sicherer, S.H.; Sampson, H.A.; Lack, G.; Beyer, K.; Oettgen, H.C. Food allergy. Nat. Rev. Dis. Prim. 2018, 4, 1–20. [Google Scholar] [CrossRef]
- Berin, M.C.; Shreffler, W.G. TH2 adjuvants: Implications for food allergy. J. Allergy Clin. Immunol. 2008, 121, 1311–1320. [Google Scholar] [CrossRef]
- Carrera, M.; Magadán, S. Proteomics for Development of Food Allergy Vaccines. Methods Mol. Biol. 2022, 2410, 673–689. [Google Scholar] [PubMed]
- Carrera, M.; Ezquerra-Brauer, J.M.; Aubourg, S.P. Characterization of the Jumbo Squid (Dosidicus gigas) Skin By-Product by Shotgun Proteomics and Protein-Based Bioinformatics. Mar. Drugs 2019, 18, 31. [Google Scholar] [CrossRef] [Green Version]
- Lozano-Ojalvo, D.; López-Fandiño, R. Immunomodulating peptides for food allergy prevention and treatment. Crit. Rev. Food Sci. Nutr. 2017, 58, 1629–1649. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; Yang, C.; Nau, F.; Pasco, M.; Juneja, L.R.; Okubo, T.; Mine, Y. Immunomodulatory effects of egg white enzymatic hydrolysates containing immunodominant epitopes in a BALB/c mouse model of egg allergy. J. Agric. Food Chem. 2009, 57, 2241–2248. [Google Scholar] [CrossRef] [PubMed]
- Rupa, P.; Mine, Y. Oral immunotherapy with immunodominant T-cell epitope peptides alleviates allergic reactions in a Balb/c mouse model of egg allergy. Allergy 2012, 67, 74–82. [Google Scholar] [CrossRef]
- Thang, C.L.; Zhao, X. Effects of orally administered immunodominant T-cell epitope peptides on cow’s milk protein allergy in a mouse model. Food Res. Int. 2015, 71, 126–131. [Google Scholar] [CrossRef]
- Tanaka, M.; Watanabe, H.; Yoshimoto, Y.; Kozai, H.; Okamoto, T. Anti-allergic effects of His-Ala-Gln tripeptide in vitro and in vivo. Biosci. Biotechnol. Biochem. 2017, 81, 380–383. [Google Scholar] [CrossRef] [PubMed]
- Rossi, M.; Ruvo, M.; Marasco, D.; Colombo, M.; Cassani, G.; Verdoliva, A. Anti-allergic properties of a new all-D synthetic immunoglobulin-binding peptide. Mol. Immunol. 2008, 45, 226–234. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.; Kim, Y.; Kim, H.Y.; Ro, J.Y.; Jeoung, D. Inhibitory mechanism of anti-allergic peptides in RBL2H3 cells. Eur. J. Pharmacol. 2008, 581, 191–203. [Google Scholar] [CrossRef]
- Verdoliva, A.; Rossi, M. IgE-Mediated Disorders: Current Therapeutics and New Strategies Involving Synthetic Peptides. Anti Inflamm. Anti Allergy Agents Med. Chem. 2008, 7, 252–263. [Google Scholar] [CrossRef]
- Vo, T.S.; Kim, Y.S.; Ngo, D.H.; Le, P.U.; Kim, S.Y.; Kim, S.K. Spirulina maxima peptides suppress mast cell degranulation via inactivating Akt and MAPKs phosphorylation in RBL-2H3 cells. Int. J. Biol. Macromol. 2018, 118, 2224–2229. [Google Scholar] [CrossRef]
- Vo, T.S.; Ngo, D.H.; Kang, K.H.; Park, S.J.; Kim, S.K. The role of peptides derived from Spirulina maxima in downregulation of FcεRI-mediated allergic responses. Mol. Nutr. Food Res. 2014, 58, 2226–2234. [Google Scholar] [CrossRef]
- Cheng, C.; Ng, D.S.W.; Chan, T.K.; Guan, S.P.; Ho, W.E.; Koh, A.H.M.; Bian, J.S.; Lau, H.Y.A.; Wong, W.S.F. Anti-allergic action of anti-malarial drug artesunate in experimental mast cell-mediated anaphylactic models. Allergy 2013, 68, 195–203. [Google Scholar] [CrossRef]
- Ko, S.C.; Lee, D.S.; Park, W.S.; Yoo, J.S.; Yim, M.J.; Qian, Z.J.; Lee, C.M.; Oh, J.; Jung, W.K.; Choi, I.W. Anti-allergic effects of a nonameric peptide isolated from the intestine gastrointestinal digests of abalone (Haliotis discus hannai) in activated HMC-1 human mast cells. Int. J. Mol. Med. 2016, 37, 243–250. [Google Scholar] [CrossRef] [Green Version]
- Wang, K.; Siddanakoppalu, P.N.; Ahmed, I.; Pavase, T.R.; Lin, H.; Li, Z. Purification and identification of anti-allergic peptide from Atlantic Salmon (Salmo salar) byproduct enzymatic hydrolysates. J. Funct. Foods 2020, 72, 104084. [Google Scholar] [CrossRef]
- Wang, K.; Pramod, S.N.; Pavase, T.R.; Ahmed, I.; Lin, H.; Liu, L.; Tian, S.; Lin, H.; Li, Z. An overview on marine anti-allergic active substances for alleviating food-induced allergy. Crit. Rev. Food Sci. Nutr. 2019, 60, 2549–2563. [Google Scholar] [CrossRef]
- Cross, M.L.; Stevenson, L.M.; Gill, H.S. Anti-allergy properties of fermented foods: An important immunoregulatory mechanism of lactic acid bacteria? Int. Immunopharmacol. 2001, 1, 891–901. [Google Scholar] [CrossRef]
- Pereyra, B.S.; Falcoff, R.; Falcoff, E.; Lemonnier, D. Interferon induction by Lactobacillus bulgaricus and Streptococcus thermophilus in mice. Eur. Cytokine Netw. 1991, 2, 299–303. [Google Scholar]
- Sütas, Y.; Hurme, M.; Isolauri, E. Down-Regulation of Anti-CD3 Antibody-Induced IL-4 Production by Bovine Caseins Hydrolysed with Lactobacillus GG-Derived Enzymes. Scand. J. Immunol. 1996, 43, 687–689. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.; Shu, Y.; Chen, J.; Cao, X. Preparation and in vitro bioactive evaluation of cashew-nut proteins hydrolysate as a potential source of anti-allergy peptides. J. Food Sci. Technol. 2021, 58, 3780–3789. [Google Scholar] [CrossRef]
- Wang, J.; Kortner, T.M.; Chikwati, E.M.; Li, Y.; Jaramillo-Torres, A.; Jakobsen, J.V.; Ravndal, J.; Brevik, Ø.J.; Einen, O.; Krogdahl, Å. Gut immune functions and health in Atlantic salmon (Salmo salar) from late freshwater stage until one year in seawater and effects of functional ingredients: A case study from a commercial sized research site in the Arctic region. Fish Shellfish Immunol. 2020, 106, 1106–1119. [Google Scholar] [CrossRef]
- Chen, L.; Deng, H.; Cui, H.; Fang, J.; Zuo, Z.; Deng, J.; Li, Y.; Wang, X.; Zhao, L. Inflammatory responses and inflammation-associated diseases in organs. Oncotarget 2018, 9, 7204. [Google Scholar] [CrossRef] [Green Version]
- Medzhitov, R. Inflammation 2010: New Adventures of an Old Flame. Cell 2010, 140, 771–776. [Google Scholar] [CrossRef] [Green Version]
- Gao, R.; Shu, W.; Shen, Y.; Sun, Q.; Bai, F.; Wang, J.; Li, D.; Li, Y.; Jin, W.; Yuan, L. Sturgeon protein-derived peptides exert anti-inflammatory effects in LPS-stimulated RAW264.7 macrophages via the MAPK pathway. J. Funct. Foods 2020, 72, 104044. [Google Scholar] [CrossRef]
- Liu, H.; Li, B. Separation and identification of collagen peptides derived from enzymatic hydrolysate of Salmo salar skin and their anti-inflammatory activity in lipopolysaccharide (LPS)-induced RAW264.7 inflammatory model. J. Food Biochem. 2022, 46, e14122. [Google Scholar] [CrossRef]
- Ahn, C.B.; Cho, Y.S.; Je, J.Y. Purification and anti-inflammatory action of tripeptide from salmon pectoral fin byproduct protein hydrolysate. Food Chem. 2015, 168, 151–156. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Sun, L.; Li, D.; Lai, X.; Wen, S.; Chen, R.; Zhang, Z.; Li, Q.; Sun, S. Green tea peptides ameliorate diabetic nephropathy by inhibiting the TGF-β/Smad signaling pathway in mice. Food Funct. 2022, 13, 3258–3270. [Google Scholar] [CrossRef] [PubMed]
- Joshi, I.; Nazeer, R.A. Anti-inflammatory potential of novel hexapeptide derived from Meretrix meretrix foot and its functional properties. Amino Acids 2020, 52, 1391–1401. [Google Scholar] [CrossRef] [PubMed]
- Liang, N.; Beverly, R.L.; Scottoline, B.P.; Dallas, D.C. Peptides Derived from In Vitro and In Vivo Digestion of Human Milk Are Immunomodulatory in THP-1 Human Macrophages. J. Nutr. 2022, 152, 331–342. [Google Scholar] [CrossRef] [PubMed]
- Jiehui, Z.; Liuliu, M.; Haihong, X.; Yang, G.; Yingkai, J.; Lun, Z.; An Li, D.X.; Dongsheng, Z.; Shaohui, Z. Immunomodulating effects of casein-derived peptides QEPVL and QEPV on lymphocytes in vitro and in vivo. Food Funct. 2014, 5, 2061–2069. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Zhang, H.; Mats, L.; Liu, R.; Deng, Z.; Mine, Y.; Tsao, R. Anti-inflammatory effect and cellular uptake mechanism of peptides from common bean (Phaseolus vulga L.) milk and yogurts in caco-2 mono-A nd caco-2/EA.hy926 co-culture models. J. Agric. Food Chem. 2019, 67, 8370–8381. [Google Scholar] [CrossRef]
- Shi, Z.; Dun, B.; Wei, Z.; Liu, C.; Tian, J.; Ren, G.; Yao, Y. Peptides Released from Extruded Adzuki Bean Protein through Simulated Gastrointestinal Digestion Exhibit Anti-inflammatory Activity. J. Agric. Food Chem. 2021, 69, 7028–7036. [Google Scholar] [CrossRef]
- Nakamura, T.; Hirota, T.; Mizushima, K.; Ohki, K.; Naito, Y.; Yamamoto, N.; Yoshikawa, T. Milk-Derived Peptides, Val-Pro-Pro and Ile-Pro-Pro, Attenuate Atherosclerosis Development in Apolipoprotein E–Deficient Mice: A Preliminary Study. J. Med. Food 2013, 16, 396–403. [Google Scholar] [CrossRef]
- Chakrabarti, S.; Wu, J. Milk-Derived Tripeptides IPP (Ile-Pro-Pro) and VPP (Val-Pro-Pro) Promote Adipocyte Differentiation and Inhibit Inflammation in 3T3-F442A Cells. PLoS ONE 2015, 10, e0117492. [Google Scholar] [CrossRef] [Green Version]
- Kennedy, K.; Keogh, B.; Lopez, C.; Adelfio, A.; Molloy, B.; Kerr, A.; Wall, A.M.; Jalowicki, G.; Holton, T.A.; Khaldi, N. An Artificial Intelligence Characterised Functional Ingredient, Derived from Rice, Inhibits TNF-α and Significantly Improves Physical Strength in an Inflammaging Population. Foods 2020, 9, 1147. [Google Scholar] [CrossRef] [PubMed]
- Vo, T.S.; Ryu, B.M.; Kim, S.K. Purification of novel anti-inflammatory peptides from enzymatic hydrolysate of the edible microalgal Spirulina maxima. J. Funct. Foods 2013, 5, 1336–1346. [Google Scholar] [CrossRef]
- Zhang, S.; Ren, M.; Zeng, X.; He, P.; Ma, X.; Qiao, S. Leucine stimulates ASCT2 amino acid transporter expression in porcine jejunal epithelial cell line (IPEC-J2) through PI3K/Akt/mTOR and ERK signaling pathways. Amino Acids 2015, 46, 2633–2642. [Google Scholar] [CrossRef]
- Singh, S.; Datta, A.; Schmidtchen, A.; Bhunia, A.; Malmsten, M. Tryptophan end-tagging for promoted lipopolysaccharide interactions and anti-inflammatory effects. Sci. Rep. 2017, 7, 212. [Google Scholar] [CrossRef]
- Gao, Y.; Qin, H.; Wu, D.; Liu, C.; Fang, L.; Wang, J.; Liu, X.; Min, W. Walnut peptide WEKPPVSH in alleviating oxidative stress and inflammation in lipopolysaccharide-activated BV-2 microglia via the Nrf2/HO-1 and NF-κB/p38 MAPK pathways. J. Biosci. Bioeng. 2021, 132, 496–504. [Google Scholar] [CrossRef] [PubMed]
- Saisavoey, T.; Sangtanoo, P.; Chanchao, C.; Reamtong, O.; Karnchanatat, A. Identification of novel anti-inflammatory peptides from bee pollen (Apis mellifera) hydrolysate in lipopolysaccharide-stimulated RAW264.7 macrophages. J. Apic. Res. 2020, 60, 280–289. [Google Scholar] [CrossRef]
- Velliquette, R.A.; Fast, D.J.; Maly, E.R.; Alashi, A.M.; Aluko, R.E. Enzymatically derived sunflower protein hydrolysate and peptides inhibit NFκB and promote monocyte differentiation to a dendritic cell phenotype. Food Chem. 2020, 319, 126563. [Google Scholar] [CrossRef]
- Feng, X.W.; Cheng, Q.L.; Fang, L.; Liu, W.Y.; Liu, L.W.; Sun, C.Q.; Lu, Z.H.; Li, G.M.; Gu, R.Z. Corn oligopeptides inhibit Akt/NF-κB signaling pathway and inflammatory factors to ameliorate CCl4-induced hepatic fibrosis in mice. J. Food Biochem. 2022, 46, e14162. [Google Scholar] [CrossRef]
- Gałązka-Czarnecka, I.; Budryn, G. Bioactive Peptide Analysis. In Analytical Methods in the Determination of Bioactive Compounds and Elements in Food; Springer: Berlin, Germany, 2021; pp. 243–262. [Google Scholar]
- Issaq, H.J.; Veenstra, T.D. Two-dimensional polyacrylamide gel electrophoresis (2D-PAGE): Advances and perspectives. Biotechniques 2008, 44, 697–700. [Google Scholar] [CrossRef] [Green Version]
- Bonfatti, V.; Grigoletto, L.; Cecchinato, A.; Gallo, L.; Carnier, P. Validation of a new reversed-phase high-performance liquid chromatography method for separation and quantification of bovine milk protein genetic variants. J. Chromatogr. A 2008, 1195, 101–106. [Google Scholar] [CrossRef]
- Agregán, R.; Echegaray, N.; López-Pedrouso, M.; Kharabsheh, R.; Franco, D.; Lorenzo, J.M. Proteomic Advances in Milk and Dairy Products. Molecules 2021, 26, 3832. [Google Scholar] [CrossRef]
- Agyei, D.; Danquah, M.K. Industrial-scale manufacturing of pharmaceutical-grade bioactive peptides. Biotechnol. Adv. 2011, 29, 272–277. [Google Scholar] [CrossRef]
- Carrasco-Castilla, J.; Hernández-Álvarez, A.J.; Jiménez-Martínez, C.; Gutiérrez-López, G.F.; Dávila-Ortiz, G. Use of Proteomics and Peptidomics Methods in Food Bioactive Peptide Science and Engineering. Food Eng. Rev. 2012, 4, 224–243. [Google Scholar] [CrossRef]
- Abd El-Fattah, A.M.; Sakr, S.S.; El-Dieb, S.M.; Elkashef, H.A.S. Bioactive peptides with ACE-I and antioxidant activity produced from milk proteolysis. Int. J. Food Prop. 2017, 20, 3033–3042. [Google Scholar] [CrossRef]
- Fu, Y.; Therkildsen, M.; Aluko, R.E.; Lametsch, R. Exploration of collagen recovered from animal by-products as a precursor of bioactive peptides: Successes and challenges. Crit. Rev. Food Sci. Nutr. 2018, 59, 2011–2027. [Google Scholar] [CrossRef]
- Wang, X.; Yu, H.; Xing, R.; Li, P. Characterization, Preparation, and Purification of Marine Bioactive Peptides. BioMed Res. Int. 2017, 2017. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- López-Expósito, I.; Gómez-Ruiz, J.Á.; Amigo, L.; Recio, I. Identification of antibacterial peptides from ovine αs2-casein. Int. Dairy J. 2006, 16, 1072–1080. [Google Scholar] [CrossRef]
- Caron, J.; Chataigné, G.; Gimeno, J.P.; Duhal, N.; Goossens, J.F.; Dhulster, P.; Cudennec, B.; Ravallec, R.; Flahaut, C. Food peptidomics of in vitro gastrointestinal digestions of partially purified bovine hemoglobin: Low-resolution versus high-resolution LC-MS/MS analyses. Electrophoresis 2016, 37, 1814–1822. [Google Scholar] [CrossRef]
- Guo, H.; Kouzuma, Y.; Yonekura, M. Structures and properties of antioxidative peptides derived from royal jelly protein. Food Chem. 2009, 113, 238–245. [Google Scholar] [CrossRef]
- Saito, K.; Jin, D.H.; Ogawa, T.; Muramoto, K.; Hatakeyama, E.; Yasuhara, T.; Nokihara, K. Antioxidative properties of tripeptide libraries prepared by the combinatorial chemistry. J. Agric. Food Chem. 2003, 51, 3668–3674. [Google Scholar] [CrossRef]
- Hernandez-Ledesma, B.; Amigo, L.; Ramos, M.; Recio, I. Angiotensin Converting Enzyme Inhibitory Activity in Commercial Fermented Products. Formation of Peptides under Simulated Gastrointestinal Digestion. J. Agric. Food Chem. 2004, 52, 1504–1510. [Google Scholar] [CrossRef]
- Weber, D.; Raymond, P.; Ben-Rejeb, S.; Lau, B. Development of a liquid chromatography-tandem mass spectrometry method using capillary liquid chromatography and nanoelectrospray ionization-quadrupole time-of-flight hybrid mass spectrometer for the detection of milk allergens. J. Agric. Food Chem. 2006, 54, 1604–1610. [Google Scholar] [CrossRef]
- Martin, M.; Wellner, A.; Ossowski, I.; Henle, T. Identification and quantification of inhibitors for angiotensin-converting enzyme in hypoallergenic infant milk formulas. J. Agric. Food Chem. 2008, 56, 6333–6338. [Google Scholar] [CrossRef]
- Català-Clariana, S.; Benavente, F.; Giménez, E.; Barbosa, J.; Sanz-Nebot, V. Identification of bioactive peptides in hypoallergenic infant milk formulas by capillary electrophoresis–mass spectrometry. Anal. Chim. Acta 2010, 683, 119–125. [Google Scholar] [CrossRef]
- Herrero, M.; Ibañez, E.; Cifuentes, A. Capillary electrophoresis-electrospray-mass spectrometry in peptide analysis and peptidomics. Electrophoresis 2008, 29, 2148–2160. [Google Scholar] [CrossRef] [Green Version]
- Staub, A.; Schappler, J.; Rudaz, S.; Veuthey, J.L. CE-TOF/MS: Fundamental concepts, instrumental considerations and applications. Electrophoresis 2009, 30, 1610–1623. [Google Scholar] [CrossRef] [PubMed]
- Aclav Kašička, V. ´; Kaši, K.; Kašička, K. Recent developments in capillary and microchip electroseparations of peptides (2011–2013). Electrophoresis 2014, 35, 69–95. [Google Scholar] [CrossRef] [PubMed]
- Gaspar, A.; Englmann, M.; Fekete, A.; Harir, M.; Schmitt-Kopplin, P. Trends in CE-MS 2005–2006. Electrophoresis 2008, 29, 66–79. [Google Scholar] [CrossRef] [PubMed]
- Muller, L.; Barták, P.; Bednář, P.; Fryšová, I.; Ševčík, J.; Lemr, K. Capillary electrophoresis-mass spectrometry—A fast and reliable tool for the monitoring of milk adulteration. Electrophoresis 2008, 29, 2088–2093. [Google Scholar] [CrossRef] [PubMed]
- Veledo, M.T.; de Frutos, M.; Diez-Masa, J.C. Analysis of trace amounts of bovine β-lactoglobulin in infant formulas by capillary electrophoresis with on-capillary derivatization and laser-induced fluorescence detection. J. Sep. Sci. 2005, 28, 941–947. [Google Scholar] [CrossRef] [PubMed]
- Ibáñez, C.; Simó, C.; García-Cañas, V.; Cifuentes, A.; Castro-Puyana, M. Metabolomics, peptidomics and proteomics applications of capillary electrophoresis-mass spectrometry in Foodomics: A review. Anal. Chim. Acta 2013, 802, 1–13. [Google Scholar] [CrossRef]
- Gómez-Ruiz, J.Á.; Leake, D.S.; Ames, J.M. In vitro antioxidant activity of coffee compounds and their metabolites. J. Agric. Food Chem. 2007, 55, 6962–6969. [Google Scholar] [CrossRef]
- Miralles, B.; Rothbauer, V.; Manso, M.A.; Amigo, L.; Krause, I.; Ramos, M. Improved method for the simultaneous determination of whey proteins, caseins and para-κ-casein in milk and dairy products by capillary electrophoresis. J. Chromatogr. A 2001, 915, 225–230. [Google Scholar] [CrossRef]
- Albillos, S.M.; Busto, M.D.; Perez-Mateos, M.; Ortega, N. Chemometrical analysis of capillary electrophoresis casein fractions for predicting ripening times of milk mixture cheese. J. Agric. Food Chem. 2005, 53, 6094–6099. [Google Scholar] [CrossRef] [PubMed]
- Molina, E.; De Frutos, M.; Ramos, M. Capillary electrophoresis characterization of the casein fraction of cheeses made from cows’, ewes’ and goats’ milks. J. Dairy Res. 2000, 67, 209–216. [Google Scholar] [CrossRef] [PubMed]
- Merz, A.; Stephan, R.; Johler, S. Staphylococcus aureus Isolates from Goat and Sheep Milk Seem to Be Closely Related and Differ from Isolates Detected from Bovine Milk. Front. Microbiol. 2016, 7, 319. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Strickland, M.; Johnson, M.E.; Broadbent, J.R. Qualitative and quantitative analysis of proteins and peptides in milk products by capillary electrophoresis-Strickland-2001-ELECTROPHORESIS-Wiley Online Library. Electrophoresis 2001, 22, 1510–1517. [Google Scholar] [CrossRef]
- Saz, J.M.; Marina, M.L. High performance liquid chromatography and capillary electrophoresis in the analysis of soybean proteins and peptides in foodstuffs. J. Sep. Sci. 2007, 30, 431–451. [Google Scholar] [CrossRef] [PubMed]
- Welch, C.J.; Wu, N.; Biba, M.; Hartman, R.; Brkovic, T.; Gong, X.; Helmy, R.; Schafer, W.; Cuff, J.; Pirzada, Z.; et al. Greening analytical chromatography. TrAC Trends Anal. Chem. 2010, 29, 667–680. [Google Scholar] [CrossRef]
- Schappler, J.; Veuthey, J.L.; Rudaz, S. 18 Coupling CE and microchip-based devices with mass spectrometry. Sep. Sci. Technol. 2008, 9, 477–521. [Google Scholar]
- Chen, C.-H.; Krupke, A.; Carrera, M.; Weisbrod, C.R.; Huguet, R.; Chen, S.-M.; Karger, A.; Williams, S.; Wenz, M.; Huhmer, A.F.; et al. Ultra-Fast Analysis of Allergens Using Capillary Electrophoresis Coupled to Mass Spectrometry and Ultra Violet Photodissociation. In Proceedings of the 15th Human Proteome Organization (HUPO) World Congress, Taipei, Taiwan, 18–22 September 2016. [Google Scholar]
- Silva, A.M.N.; Vitorino, R.; Domingues, M.R.M.; Spickett, C.M.; Domingues, P. Post-translational Modifications and Mass Spectrometry Detection. Free Radic. Biol. Med. 2013, 65, 925–941. [Google Scholar] [CrossRef] [Green Version]
- Panchaud, A.; Affolter, M.; Kussmann, M. Mass spectrometry for nutritional peptidomics: How to analyze food bioactives and their health effects. J. Proteomics 2012, 75, 3546–3559. [Google Scholar] [CrossRef]
- Gómez-Ruiz, J.Á.; Taborda, G.; Amigo, L.; Recio, I.; Ramos, M. Identification of ACE-inhibitory peptides in different Spanish cheeses by tandem mass spectrometry. Eur. Food Res. Technol. 2006, 223, 595–601. [Google Scholar] [CrossRef]
- Leopold, J.; Popkova, Y.; Engel, K.M.; Schiller, J. Recent Developments of Useful MALDI Matrices for the Mass Spectrometric Characterization of Lipids. Biomolecules 2018, 8, 173. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ayala-Niño, A.; Rodríguez-Serrano, G.M.; González-Olivares, L.G.; Contreras-López, E.; Regal-López, P.; Cepeda-Saez, A. Sequence Identification of Bioactive Peptides from Amaranth Seed Proteins (Amaranthus hypochondriacus spp.). Molecules 2019, 24, 3033. [Google Scholar] [CrossRef]
- Silva-Sánchez, C.; Barba De La Rosa, A.P.; León-Galván, M.F.; De Lumen, B.O.; De León-Rodríguez, A.; González De Mejía, E. Bioactive peptides in amaranth (Amaranthus hypochondriacus) seed. J. Agric. Food Chem. 2008, 56, 1233–1240. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Dia, V.P.; Vasconez, M.; De Mejia, E.G.; Nelson, R.L. Analysis of Soybean Protein-Derived Peptides and the Effect of Cultivar, Environmental Conditions, and Processing on Lunasin Concentration in Soybean and Soy Products. J. AOAC Int. 2008, 91, 936–946. [Google Scholar] [CrossRef] [Green Version]
- Ma, B.; Johnson, R. De Novo Sequencing and Homology Searching. Mol. Cell. Proteom. 2012, 11, O111.014902. [Google Scholar] [CrossRef] [Green Version]
- Picariello, G.; Mamone, G.; Nitride, C.; Addeo, F.; Ferranti, P. Protein digestomics: Integrated platforms to study food-protein digestion and derived functional and active peptides. TrAC Trends Anal. Chem. 2013, 52, 120–134. [Google Scholar] [CrossRef]
- Kastin, A. Handbook of Biologically Active Peptides, 2nd ed.; Elsevier: Amsterdam, The Netherlands, 2013; ISBN 9780123850966. [Google Scholar]
- Fekete, S.; Veuthey, J.L.; Guillarme, D. New trends in reversed-phase liquid chromatographic separations of therapeutic peptides and proteins: Theory and applications. J. Pharm. Biomed. Anal. 2012, 69, 9–27. [Google Scholar] [CrossRef]
- Boutrou, R.; Gaudichon, C.; Dupont, D.; Jardin, J.; Airinei, G.; Marsset-Baglieri, A.; Benamouzig, R.; Tomé, D.; Leonil, J. Sequential release of milk protein–derived bioactive peptides in the jejunum in healthy humans. Am. J. Clin. Nutr. 2013, 97, 1314–1323. [Google Scholar] [CrossRef] [Green Version]
- Shazly, A.B.; He, Z.; El-Aziz, M.A.; Zeng, M.; Zhang, S.; Qin, F.; Chen, J. Fractionation and identification of novel antioxidant peptides from buffalo and bovine casein hydrolysates. Food Chem. 2017, 232, 753–762. [Google Scholar] [CrossRef]
- Peighambardoust, S.H.; Karami, Z.; Pateiro, M.; Lorenzo, J.M. A Review on Health-Promoting, Biological, and Functional Aspects of Bioactive Peptides in Food Applications. Biomolecules 2021, 11, 631. [Google Scholar] [CrossRef]
- Fideler, J.; Johanningsmeier, S.D.; Ekelöf, M.; Muddiman, D.C. Discovery and quantification of bioactive peptides in fermented cucumber by direct analysis IR-MALDESI mass spectrometry and LC-QQQ-MS. Food Chem. 2019, 271, 715–723. [Google Scholar] [CrossRef] [PubMed]
- López-Pedrouso, M.; Borrajo, P.; Pateiro, M.; Lorenzo, J.M.; Franco, D. Antioxidant activity and peptidomic analysis of porcine liver hydrolysates using alcalase, bromelain, flavourzyme and papain enzymes. Food Res. Int. 2020, 137, 109389. [Google Scholar] [CrossRef] [PubMed]
- Mahdi, C.; Untari, H.; Padaga, M.C. Identification and Characterization of Bioactive Peptides of Fermented Goat Milk as a Sources of Antioxidant as a Therapeutic Natural Product. IOP Conf. Ser. Mater. Sci. Eng. 2018, 299, 012014. [Google Scholar] [CrossRef]
- Wang, L.; Ma, M.; Yu, Z.; Du, S. kui Preparation and identification of antioxidant peptides from cottonseed proteins. Food Chem. 2021, 352, 129399. [Google Scholar] [CrossRef]
- Chai, T.T.; Xiao, J.; Mohana Dass, S.; Teoh, J.Y.; Ee, K.Y.; Ng, W.J.; Wong, F.C. Identification of antioxidant peptides derived from tropical jackfruit seed and investigation of the stability profiles. Food Chem. 2021, 340, 127876. [Google Scholar] [CrossRef]
- Wu, D.; Li, M.; Ding, J.; Zheng, J.; Zhu, B.W.; Lin, S. Structure-activity relationship and pathway of antioxidant shrimp peptides in a PC12 cell model. J. Funct. Foods 2020, 70, 103978. [Google Scholar] [CrossRef]
- Carrera, M.; Cañas, B.; Gallardo, J.M. The sarcoplasmic fish proteome: Pathways, metabolic networks and potential bioactive peptides for nutritional inferences. J. Proteomics 2013, 78, 211–220. [Google Scholar] [CrossRef]
- Peng, L.; Kong, X.; Wang, Z.; Ai-lati, A.; Ji, Z.; Mao, J. Baijiu vinasse as a new source of bioactive peptides with antioxidant and anti-inflammatory activity. Food Chem. 2021, 339, 128159. [Google Scholar] [CrossRef]
- Gao, R.; Shu, W.; Shen, Y.; Sun, Q.; Jin, W.; Li, D.; Li, Y.; Yuan, L. Peptide fraction from sturgeon muscle by pepsin hydrolysis exerts anti-inflammatory effects in LPS-stimulated RAW264.7 macrophages via MAPK and NF-κB pathways. Food Sci. Hum. Wellness 2021, 10, 103–111. [Google Scholar] [CrossRef]
- Sandoval-Sicairos, E.S.; Milán-Noris, A.K.; Luna-Vital, D.A.; Milán-Carrillo, J.; Montoya-Rodríguez, A. Anti-inflammatory and antioxidant effects of peptides released from germinated amaranth during in vitro simulated gastrointestinal digestion. Food Chem. 2021, 343, 128394. [Google Scholar] [CrossRef] [PubMed]
- Heymich, M.L.; Friedlein, U.; Trollmann, M.; Schwaiger, K.; Böckmann, R.A.; Pischetsrieder, M. Generation of antimicrobial peptides Leg1 and Leg2 from chickpea storage protein, active against food spoilage bacteria and foodborne pathogens. Food Chem. 2021, 347, 128917. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Kang, H.K.; Cheong, H.; Park, Y. A Novel Antimicrobial Peptides From Pine Needles of Pinus densiflora Sieb. et Zucc. Against Foodborne Bacteria. Front. Microbiol. 2021, 12, 1129. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Zhang, M.; Pan, F.; Li, J.; Dou, R.; Wang, X.; Wang, Y.; He, Y.; Wang, S.; Cai, S. In silico analysis of novel dipeptidyl peptidase-IV inhibitory peptides released from Macadamia integrifolia antimicrobial protein 2 (MiAMP2) and the possible pathways involved in diabetes protection. Curr. Res. Food Sci. 2021, 4, 603–611. [Google Scholar] [CrossRef]
- Khani, S.; Seyedjavadi, S.S.; Zare-Zardini, H.; Hosseini, H.M.; Goudarzi, M.; Khatami, S.; Amani, J.; Imani Fooladi, A.A.; Razzaghi-Abyaneh, M. Isolation and functional characterization of an antifungal hydrophilic peptide, Skh-AMP1, derived from Satureja khuzistanica leaves. Phytochemistry 2019, 164, 136–143. [Google Scholar] [CrossRef]
- Aiemratchanee, P.; Panyawechamontri, K.; Phaophu, P.; Reamtong, O.; Panbangred, W. In vitro antihypertensive activity of bioactive peptides derived from porcine blood corpuscle and plasma proteins. Int. J. Food Sci. Technol. 2021, 56, 2315–2324. [Google Scholar] [CrossRef]
- Choe, J.; Seol, K.H.; Kim, H.J.; Hwang, J.T.; Lee, M.; Jo, C. Isolation and identification of angiotensin I-converting enzyme inhibitory peptides derived from thermolysin-injected beef M. longissimus. Asian-Australas. J. Anim. Sci. 2019, 32, 430. [Google Scholar] [CrossRef] [Green Version]
- Karami, Z.; Akbari-adergani, B. Bioactive food derived peptides: A review on correlation between structure of bioactive peptides and their functional properties. J. Food Sci. Technol. 2019, 56, 535–547. [Google Scholar] [CrossRef]
- Amado, I.R.; Vázquez, J.A.; Gonzaĺez, P.; Esteban-Fernańdez, D.; Carrera, M.; Piñeiro, C. Identification of the Major ACE-Inhibitory Peptides Produced by Enzymatic Hydrolysis of a Protein Concentrate from Cuttlefish Wastewater. Mar. Drugs 2014, 12, 1390–1405. [Google Scholar] [CrossRef] [Green Version]
- Mudgil, P.; Kamal, H.; Priya Kilari, B.; Mohd Salim, M.A.S.; Gan, C.Y.; Maqsood, S. Simulated gastrointestinal digestion of camel and bovine casein hydrolysates: Identification and characterization of novel anti-diabetic bioactive peptides. Food Chem. 2021, 353, 129374. [Google Scholar] [CrossRef]
- Baba, W.N.; Mudgil, P.; Kamal, H.; Kilari, B.P.; Gan, C.Y.; Maqsood, S. Identification and characterization of novel α-amylase and α-glucosidase inhibitory peptides from camel whey proteins. J. Dairy Sci. 2021, 104, 1364–1377. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Wu, T.; Fang, L.; Liu, C.; Liu, X.; Li, H.; Shi, J.; Li, M.; Min, W. Anti-diabetic effect by walnut (Juglans mandshurica Maxim.)-derived peptide LPLLR through inhibiting α-glucosidase and α-amylase, and alleviating insulin resistance of hepatic HepG2 cells. J. Funct. Foods 2020, 69, 103944. [Google Scholar] [CrossRef]
- Siow, H.L.; Lim, T.S.; Gan, C.Y. Development of a workflow for screening and identification of α-amylase inhibitory peptides from food source using an integrated Bioinformatics-phage display approach: Case study–Cumin seed. Food Chem. 2017, 214, 67–76. [Google Scholar] [CrossRef] [PubMed]
- Quintero-Soto, M.F.; Chávez-Ontiveros, J.; Garzón-Tiznado, J.A.; Salazar-Salas, N.Y.; Pineda-Hidalgo, K.V.; Delgado-Vargas, F.; López-Valenzuela, J.A. Characterization of peptides with antioxidant activity and antidiabetic potential obtained from chickpea (Cicer arietinum L.) protein hydrolyzates. J. Food Sci. 2021, 86, 2962–2977. [Google Scholar] [CrossRef] [PubMed]
- Bravo, F.I.; Mas-Capdevila, A.; López-Fernández-Sobrino, R.; Torres-Fuentes, C.; Mulero, M.; Alcaide-Hidalgo, J.M.; Muguerza, B. Identification of novel antihypertensive peptides from wine lees hydrolysate. Food Chem. 2022, 366, 130690. [Google Scholar] [CrossRef]
- Song, Y.; Gu, H.; Jo, J.M.; Shin, M.; Kim, S.Y.; Gam, D.H.; Imamura, S.; Kim, J.W. Production of Functional Peptide with Anti-obesity Effect from Defatted Tenebrio molitor Larvae Using Proteolytic Enzyme. Biotechnol. Bioprocess Eng. 2020, 25, 374–383. [Google Scholar] [CrossRef]
- FAO. The State of World Fisheries and Aquaculture. 2020. Available online: https://www.fao.org/documents/card/en/c/ca9229en (accessed on 8 September 2022).
- Xu, Y.; Chen, C.; Ji, D.; Hang, N.; Xie, C. Proteomic profile analysis of Pyropia haitanensis in response to high-temperature stress. J. Appl. Phycol. 2014, 26, 607–618. [Google Scholar] [CrossRef]
- López-Cristoffanini, C.; Zapata, J.; Gaillard, F.; Potin, P.; Correa, J.A.; Contreras-Porcia, L. Identification of proteins involved in desiccation tolerance in the red seaweed Pyropia orbicularis (Rhodophyta, Bangiales). Proteomics 2015, 15, 3954–3968. [Google Scholar] [CrossRef]
- Du, H.; Liang, H.; Jiang, Y.; Qu, X.; Yan, H.; Liu, X. Proteome responses of Gracilaria lemaneiformis exposed to lead stress. Mar. Pollut. Bull. 2018, 135, 311–317. [Google Scholar] [CrossRef]
- Deng, Y.; Yao, J.; Wang, X.; Guo, H.; Duan, D. Transcriptome Sequencing and Comparative Analysis of Saccharina japonica (Laminariales, Phaeophyceae) under Blue Light Induction. PLoS ONE 2012, 7, e39704. [Google Scholar] [CrossRef]
- Ritter, A.; Dittami, S.M.; Goulitquer, S.; Correa, J.A.; Boyen, C.; Potin, P.; Tonon, T. Transcriptomic and metabolomic analysis of copper stress acclimation in Ectocarpus siliculosus highlights signaling and tolerance mechanisms in brown algae. BMC Plant Biol. 2014, 14, 1–17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Dong, C.; Xue, Z.; Jin, Q.; Xu, Y. De novo transcriptome sequencing and discovery of genes related to copper tolerance in Paeonia ostii. Gene 2016, 576, 126–135. [Google Scholar] [CrossRef] [PubMed]
- Gao, M.; Zhang, S.; Luo, C.; He, X.; Wei, S.; Jiang, W.; He, F.; Lin, Z.; Yan, M.; Dong, W. Transcriptome analysis of starch and sucrose metabolism across bulb development in Sagittaria sagittifolia. Gene 2018, 649, 99–112. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, X.; Shan, T.; Pang, S.; Xu, N. Transcriptome profiling of the meristem tissue of Saccharina japonica (Phaeophyceae, Laminariales) under severe stress of copper. Mar. Genom. 2019, 47, 100671. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Cao, Z.; Liang, G.; Li, X.; Wu, H.; Yang, G. Comparative Transcriptome Analysis Reveals Candidate Genes Related to Structural and Storage Carbohydrate Biosynthesis in Kelp Saccharina japonica (Laminariales, Phaeophyceae). J. Phycol. 2020, 56, 1168–1183. [Google Scholar] [CrossRef] [PubMed]
- Carrera, M.; Pazos, M.; Aubourg, S.P.; Gallardo, J.M. Shotgun Proteomics and Protein-Based Bioinformatics for the Characterization of Food-Derived Bioactive Peptides. Methods Mol. Biol. 2021, 2259, 215–223. [Google Scholar] [PubMed]
- Sanger, F. The Arrangement of Amino Acids in Proteins. Adv. Protein Chem. 1952, 7, 1–67. [Google Scholar] [PubMed]
- Agyei, D.; Tsopmo, A.; Udenigwe, C.C. Bioinformatics and peptidomics approaches to the discovery and analysis of food-derived bioactive peptides. Anal. Bioanal. Chem. 2018, 410, 3463–3472. [Google Scholar] [CrossRef]
- Minkiewicz, P.; Dziuba, J.; Darewicz, M.; Iwaniak, A.; Michalska, J. Online Programs and Databases of Peptides and Proteolytic Enzymes–A Brief Update for 2007–2008. Food Technol. Biotechnol. 2009, 47, 345–355. [Google Scholar]
- Minkiewicz, P.; Dziuba, J.; Iwaniak, A.; Dziuba, M.; Darewicz, M. BIOPEP Database and Other Programs for Processing Bioactive Peptide Sequences. J. AOAC Int. 2008, 91, 965–980. [Google Scholar] [CrossRef] [Green Version]
- Minkiewicz, P.; Dziuba, J.; Gorzata Darewicz, M.; Iwaniak, A.; Dziuba, M.; Nałeçz, D. Food Peptidomics. Food Technol. Biotechnol. 2008, 46, 1–10. [Google Scholar]
- Singh, S.; Singh, H.; Tuknait, A.; Chaudhary, K.; Singh, B.; Kumaran, S.; Raghava, G.P.S. PEPstrMOD: Structure prediction of peptides containing natural, non-natural and modified residues. Biol. Direct 2015, 10, 1–19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sánchez-Rivera, L.; Martínez-Maqueda, D.; Cruz-Huerta, E.; Miralles, B.; Recio, I. Peptidomics for discovery, bioavailability and monitoring of dairy bioactive peptides. Food Res. Int. 2014, 63, 170–181. [Google Scholar] [CrossRef]
- Gu, Y.; Majumder, K.; Wu, J. QSAR-aided in silico approach in evaluation of food proteins as precursors of ACE inhibitory peptides. Food Res. Int. 2011, 44, 2465–2474. [Google Scholar] [CrossRef]
- Wu, J.; Aluko, R.E.; Nakai, S. Structural requirements of angiotensin I-converting enzyme inhibitory peptides: Quantitative structure-activity relationship study of Di- and tripeptides. J. Agric. Food Chem. 2006, 54, 732–738. [Google Scholar] [CrossRef] [PubMed]
- Barkia, A.; Bougatef, A.; Khaled, H.B.; Nasri, M. Antioxidant Activities of Sardinelle HEADS and/or Viscera Protein Hydrolysates Prepared by Enzymatic Treatment. J. Food Biochem. 2010, 34, 303–320. [Google Scholar] [CrossRef]
- Meng, X.-Y.; Zhang, H.-X.; Mezei, M.; Cui, M. Molecular Docking: A Powerful Approach for Structure-Based Drug Discovery. Curr. Comput. Aided-Drug Des. 2012, 7, 146–157. [Google Scholar] [CrossRef]
- Chamata, Y.; Watson, K.A.; Jauregi, P. Whey-Derived Peptides Interactions with ACE by Molecular Docking as a Potential Predictive Tool of Natural ACE Inhibitors. Int. J. Mol. Sci. 2020, 21, 864. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Udenigwe, C.C.; Okolie, C.L.; Qian, H.; Ohanenye, I.C.; Agyei, D.; Aluko, R.E. Ribulose-1,5-bisphosphate carboxylase as a sustainable and promising plant source of bioactive peptides for food applications. Trends Food Sci. Technol. 2017, 69, 74–82. [Google Scholar] [CrossRef] [Green Version]
- Udenigwe, C.C.; Gong, M.; Wu, S. In silico analysis of the large and small subunits of cereal RuBisCO as precursors of cryptic bioactive peptides. Process Biochem. 2013, 48, 1794–1799. [Google Scholar] [CrossRef]
- Gu, Y.; Wu, J. LC–MS/MS coupled with QSAR modeling in characterising of angiotensin I-converting enzyme inhibitory peptides from soybean proteins. Food Chem. 2013, 141, 2682–2690. [Google Scholar] [CrossRef] [PubMed]
- Majumder, K.; Wu, J. A new approach for identification of novel antihypertensive peptides from egg proteins by QSAR and bioinformatics. Food Res. Int. 2010, 43, 1371–1378. [Google Scholar] [CrossRef]
- Nongonierma, A.B.; Fitzgerald, R.J. Structure activity relationship modelling of milk protein-derived peptides with dipeptidyl peptidase IV (DPP-IV) inhibitory activity. Peptides 2016, 79, 1–7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sagardia, I.; Iloro, I.; Elortza, F.; Bald, C. Quantitative structure–activity relationship based screening of bioactive peptides identified in ripened cheese. Int. Dairy J. 2013, 33, 184–190. [Google Scholar] [CrossRef]
- Keska, P.; Stadnik, J. Antimicrobial Peptides of Meat Origin-An In silico and In vitro Analysis. Protein Pept. Lett. 2017, 24, 165–173. [Google Scholar] [CrossRef]
- Lafarga, T.; O’Connor, P.; Hayes, M. Identification of novel dipeptidyl peptidase-IV and angiotensin-I-converting enzyme inhibitory peptides from meat proteins using in silico analysis. Peptides 2014, 59, 53–62. [Google Scholar] [CrossRef]
- Lafarga, T.; O’Connor, P.; Hayes, M. In silico methods to identify meat-derived prolyl endopeptidase inhibitors. Food Chem. 2015, 175, 337–343. [Google Scholar] [CrossRef]
- Martínez-Villaluenga, C.; Hernández-Ledesma, B. Peptides for Health Benefits 2019. Int. J. Mol. Sci. 2020, 21, 2543. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tejano, L.A.; Peralta, J.P.; Yap, E.E.S.; Panjaitan, F.C.A.; Chang, Y.W. Prediction of Bioactive Peptides from Chlorella sorokiniana Proteins Using Proteomic Techniques in Combination with Bioinformatics Analyses. Int. J. Mol. Sci. 2019, 20, 1786. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, B.B.; Lin, H.C.; Chang, Y.W. Analysis of proteins and potential bioactive peptides from tilapia (Oreochromis spp.) processing co-products using proteomic techniques coupled with BIOPEP database. J. Funct. Foods 2015, 19, 629–640. [Google Scholar] [CrossRef]
- Chen, J.; Sun, S.; Li, Y.; Liu, R. Proteolysis of tilapia skin collagen: Identification and release behavior of ACE-inhibitory peptides. LWT 2021, 139, 110502. [Google Scholar] [CrossRef]
- Panjaitan, F.C.A.; Gomez, H.L.R.; Chang, Y.W. In Silico Analysis of Bioactive Peptides Released from Giant Grouper (Epinephelus lanceolatus) Roe Proteins Identified by Proteomics Approach. Molecules 2018, 23, 2910. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Patil, P.J.; Usman, M.; Zhang, C.; Mehmood, A.; Zhou, M.; Teng, C.; Li, X. An updated review on food-derived bioactive peptides: Focus on the regulatory requirements, safety, and bioavailability. Compr. Rev. Food Sci. Food Saf. 2022, 21, 1732–1776. [Google Scholar] [CrossRef] [PubMed]



| Source | Peptide | Activity |
|---|---|---|
| Lactobacillus rhamnosus, plus digestion with pepsin | DKIHPFQEPVL | ACE b inhibitor |
| Lactobacillus helveticus | VPPIPP | ACE inhibitor |
| Lactobacillus GG plus pepsin and trypsin | YPFPAVPYPQRTTMPLW | Opioid, ACE inhibitor, immune-stimulator |
| Lactobacillus delbrueckii subsp., bulgaricus IFO13953 | ARHPHPLSFM | Antioxidant |
| Kluyveromyces marxianus | Tyr-Leu-Leu-Phe YLLF | ACE inhibitor |
| β-Casein-derived peptides | Lys-Val-Leu-Pro-Val-P(Glu) KVLPVP(E) | ACE inhibitor |
| Source/Allergen | Peptide Sequence | Activity | Reference |
|---|---|---|---|
| Eggs/Ovalbumin (OVA) | - AMVYLGAKDSTRTQ - SWVESQTNGIIRNVL - AAHAEINEAGREVVG | ↓Symptoms ↓Histamine ↓OVA-specific IgE ↑OVA-specific fecal IgA | [39] |
| Eggs/Ovomucoid (OVM) | - DNKTYGNKSNFSNAV | ↓Symptoms ↓Histamine ↓OVM-specific IgE, ↑IgG1, ↓IgG2a, ↑fecal IgA ↓IL-4, ↑IL-12, ↑IL-10 released by OVM splenocytes | [40] |
| Milk/β-lactoglobulin (BLG) | - AQKKIIAEKTKIPAVFKIDALN - ALKALPMHIRLSFNP | ↓Symptoms and temperature No change in BLG-specific IgE, IgG1, IgG2 or fecal IgA ↑IFN-γ, ↑IL-12, ↑IL-10 released by BLG splenocytes | [41] |
| Milk/Casein | - HAQ | ↓inflammatory cytokines (IL-4) ↓Anaphylaxis-like symptoms | [42] |
| Pooled sera of allergen patients | - LSYLLWRSRLP - LVAHVGAGGVL - RVSSCRGRNHIV - ETIGARWVRIE - TDGVTYTNDCL - RVVRYDADFWI - GFWCRRSGLVGV | ↓histamine, ↓calcium influx, ↓β- hexosaminidase, ↓phosphorylation of extracellular regulated kinase (ERK) | [43,44] |
| Synthesis | - RTY | ↓mast cell degranulation and release of β-hexosaminidase | [43,45] |
| Spirulina maxima | - LDAVNR - MMLDF - ADSDGK | ↓Histamine ↓intracelular Ca2+ | [46,47,48] |
| Mollusk/Abalone intestine | - PFNQGTFAS | ↓histamine, ↓PCA ↓inflammatory cytokines (TNF-α, IL-1β and IL-6) | [49] |
| Fish/Atlantic salmon byproduct | - TPEVHIAVDKF | ↓β-hexosaminidase for IgE-mediated RBL-2H3 cell | [50,51] |
| Source | Peptide Sequence | Model | Activity | Reference |
|---|---|---|---|---|
| Sturgeon muscle | - KIWHHTF - VHYAGTVDY - HLDDALRGQE | LPS-stimulated RAW264.7 cells | ↓MAPK pathway ↓inflammatory mediators (NO, IL-6, and IL-1β) ↑SOD activity ↓MAPKS phosphorylation | [59] |
| Salmon salar skin | - APD - QA - KA - WG | Macrophages from RAW264.7 cells | ↓NO, IL-6, IL-1β, and TNF-α | [60] |
| Salmon pectoral fit | -PAY | LPS-stimulated RAW264.7 cells | ↓NO and PGE2 ↓inflammatory cytokines (TNF-α, IL6 and IL1β) | [61] |
| Juice of cooked tuna | PRRTRMMNGGR | LPS-stimulated RAW264.7 cells | ↓inflammatory cytokines TNF-α, IFN-γ, and IL-2 | [62] |
| Meretrix meretrix clams | NPAQDC | Macrophages from RAW264.7 cells | ↓(COX)-2 activation ↓Pro-inflammatory cytokines ↓NO production | [63] |
| In-vitro-digested human milk and pooled intestinal samples from 8 infants fed human milk | 13 peptides | LPS-treated human immune THP-1 macrophages | ↓TNF-α and IL-8 | [64] |
| Milk casein | QEPVL | Lymphocytes from male Balb/c mice | ↓NO production ↓cytokines IL-4, IL-10, IFN-γ, and TNF-α | [65] |
| Gastrointestinal digestates of common bean milk and yogurt | γ-E-S-(Me)C γ-EL LLV | Basolateral EA.hy926 cells as shown cascades | ↓TNF-α induced pro-inflammatory mediators of the nuclear factor κB (NF-κB) and mitogen-activated protein kinase (MAPK) signal | [66] |
| Simulated gastrointestinal digestion of extruded adzuki bean protein | KQSESHFVDAQPEQQQR | LPS-induced RAW 264.7 macrophages | ↓production of IL-1, IL-6, TNF-α, and MCP-1. | [67] |
| Milk casein | VPP IPP | ApoE knockout mice | ↓production of IL-6, IL-1β ↓expression of NF-κB-related genes, CD40, LCK, PIK3CG, IL1B, and MAP2K7 | [68] |
| Milk casein | VPP IPP | VPP Murine preadipocyte cell line 3T3-F442A | ↑Upregulated PPARg and adiponectin expression; ↓adipokine levels and NF-κB pathway | [69] |
| Category | Name | Website | Function |
|---|---|---|---|
| Protein database | NCBI Protein database | https://www.ncbi.nlm.nih.gov/ | Basic sequence information for proteins |
| UniProtKB | http://www.uniprot.org/ | Basic sequence and structural information for proteins | |
| BIOPEP | http://www.uwm.edu.pl/biochemia/index.php/en/biopep | Protein sequence database | |
| RCSB Protein Data Bank | https://www.rcsb.org/pdb/home/home.do | ||
| PepBank | http://pepbank.mgh.harvard.edu/ | ||
| MilkAMP | http://milkampdb.org/ | ||
| PeptideDB | http://www.peptides.be/ | ||
| AMPer | http://marray.cmdr.ubc.ca/cgi-bin/amp.pl | ||
| BioPD | http://biopd.bjmu.edu.cn/ | ||
| SwePep | http://www.swepep.org/ | ||
| EROP-Moscow | http://erop.inbi.ras.ru/ | ||
| In silico digestion tools | Peptide Cutter | http://web.expasy.org/peptide_cutter/ | Server for predicting potential cleavage sites cleaved by proteases or chemicals in a given protein sequence |
| BIOPEP | http://www.uwm.edu.pl/biochemia/index.php/en/biopep | Server for predicting potential cleavage sites cleaved by proteases in a given protein sequence | |
| Enzyme Predictor | http://bioware.ucd.ie/~enzpred/Enzpred.php | Tool to evaluate the evidence for which enzymes are most likely to have cleaved a sample containing peptides from hydrolyzed proteins | |
| Bioactive peptide database BIOPEP | http://www.uwm.edu.pl/biochemia/index.php/en/biopep | Bioactive peptide database | |
| BitterDB | http://bitterdb.agri.huji.ac.il/bitterdb/ | Bitter compounds database | |
| EROP-Moscow database | http://erop.inbi.ras.ru | Database of biologically active peptides | |
| APD | http://aps.unmc.edu/AP/main.html | Several types of bioactive peptide databases with the main focus on antimicrobial peptides | |
| PeptideDB | http://www.peptides.be/ | Biologically active peptide database | |
| PepBank | http://pepbank.mgh.harvard.edu/ | Biologically active peptide database providing a search program for fragments with sequence similar to the peptides in thedatabase | |
| POPS | http://pops.csse.monash.edu.au/pops-cgi/index.php | ||
| AHTPDB | http://crdd.osdd.net/raghava/ahtpdb/ | Antihypertensive peptide database | |
| Potential bioactivity prediction | BIOPEP | http://www.uwm.edu.pl/biochemia/index.php/en/biopep | Tool for the evaluation of proteins as the precursors of bioactive peptides |
| PeptideRanker | http://bioware.ucd.ie/~compass/biowareweb/Server_pages/peptideranker.php | Server for the prediction of bioactive peptides. | |
| PeptideLocator | http://bioware.ucd.ie/ | ||
| AntiBP2 | http://crdd.osdd.net/raghava//antibp2/ | Predicting the antibacterial peptides in a protein sequence | |
| Allergenicity/toxicity prediction/analyzing | AlgPred | http://crdd.osdd.net/raghava//algpred/ | Predicting allergenic proteins and peptides |
| BIOPEP | http://www.uwm.edu.pl/biochemia/index.php/en/biopep | Allergenic protein database | |
| ToxinPred | http://crdd.osdd.net/raghava//toxinpred/ | Predicting toxicity of peptides | |
| Physicochemical characteristics prediction | Expasy-Compute pI/Mw | http://web.expasy.org/compute_pi/ | Tool to compute the theoretical pI (isoelectric point) and Mw (molecular weight) |
| ProtParam | http://web.expasy.org/protparam/ | Tool to compute grand average of hydropathicity (GRAVY) and instability index | |
| PepDraw | http://www.tulane.edu/~biochem/WW/PepDraw/ | Tool to compute net charge and hydrophobicity | |
| Peptide Structure Prediction | Server Pep-Fold | http://bioserv.rpbs.univ-paris-diderot.fr/services/PEP-FOLD/ | Tool to predict peptide structures from amino acid sequences |
| PEPstrMOD | http://osddlinux.osdd.net/raghava/pepstrmod | Server to predict the tertiary structure of small peptides | |
| Protein Structure Prediction | Server I-TASSER | https://zhanglab.ccmb.med.umich.edu/I-TASSER/ | Protein structure and function prediction |
| Mainly Software for the study of protein—ligand interactions | Discovery studio | ||
| Sybly | |||
| Autodock vina | |||
| Schrodinger | |||
| Dock | |||
| FlexX | |||
| ICM-Docking | |||
| GOLD | |||
| I-TASSER | https://zhanglab.ccmb.med.umich.edu/I-TASSER/ | Protein structure and function prediction | |
| In silico tools for molecular docking of peptides | DOCK Blaster | http://blaster.docking.org/ | |
| 1-CLICK DOCKING | https://mcule.com/apps/1-click-docking/ | ||
| BSP-SLIM | https://zhanglab.ccmb.med.umich.edu/BSP-SLIM/ | ||
| SwissDock | http://www.swissdock.ch/ | ||
| FlexPepDock | http://flexpepdock.furmanlab.cs.huji.ac.il/ | ||
| Identification and characterization of peptides, including tools for chemometrics | PubChem | https://pubchem.ncbi.nlm.nih.gov/ | |
| ProtParam | https://web.expasy.org/protparam/ | ||
| FooDB | http://foodb.ca/ | ||
| Chemical Entities of Biological Interest (ChEBI) | https://www.ebi.ac.uk/chebi/ | ||
| AAindex | http://www.genome.jp/aaindex/ | ||
| Human Metabolome Database (HMDB) | http://www.hmdb.ca/ | ||
| Peptigram | http://bioware.ucd.ie/peptigram/ | ||
| METLIN | https://metlin.scripps.edu/ | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Abril, A.G.; Pazos, M.; Villa, T.G.; Calo-Mata, P.; Barros-Velázquez, J.; Carrera, M. Proteomics Characterization of Food-Derived Bioactive Peptides with Anti-Allergic and Anti-Inflammatory Properties. Nutrients 2022, 14, 4400. https://doi.org/10.3390/nu14204400
Abril AG, Pazos M, Villa TG, Calo-Mata P, Barros-Velázquez J, Carrera M. Proteomics Characterization of Food-Derived Bioactive Peptides with Anti-Allergic and Anti-Inflammatory Properties. Nutrients. 2022; 14(20):4400. https://doi.org/10.3390/nu14204400
Chicago/Turabian StyleAbril, Ana G., Manuel Pazos, Tomás G. Villa, Pilar Calo-Mata, Jorge Barros-Velázquez, and Mónica Carrera. 2022. "Proteomics Characterization of Food-Derived Bioactive Peptides with Anti-Allergic and Anti-Inflammatory Properties" Nutrients 14, no. 20: 4400. https://doi.org/10.3390/nu14204400
APA StyleAbril, A. G., Pazos, M., Villa, T. G., Calo-Mata, P., Barros-Velázquez, J., & Carrera, M. (2022). Proteomics Characterization of Food-Derived Bioactive Peptides with Anti-Allergic and Anti-Inflammatory Properties. Nutrients, 14(20), 4400. https://doi.org/10.3390/nu14204400