Co-Cultures of Lactobacillus acidophilus and Bacillus subtilis Enhance Mucosal Barrier by Modulating Gut Microbiota-Derived Short-Chain Fatty Acids
Abstract
:1. Introduction
2. Materials and Methods
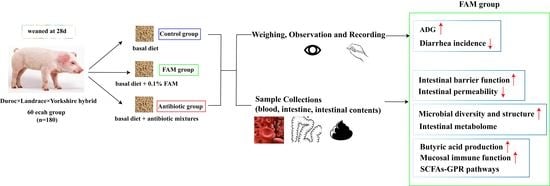
2.1. Animals and Experimental Design
2.2. Sample Collection
2.3. Intestinal Histomorphology
2.4. Gene Expression Determined by Real-Time Quantitative PCR (qRT-PCR)
2.5. Western Blot (WB)
2.6. Enzyme-Linked Immunosorbent Assay (ELISA)
2.7. Immunohistochemistry
2.8. Colonic Content Microbiome Analysis
2.9. Untargeted Metabolomic Analysis
2.10. Targeted Metabonomic Analysis
2.11. Statistical Analysis
3. Results
3.1. FAM Supplementation Promoted Intestinal Barrier Function
3.2. FAM Regulated Microbial Diversity and Structure in the Colon
3.3. Functional Metagenomics Prediction and Spearman’s Correlation Analysis of Gut Microbiota
3.4. FAM Induced Functional Changes of Intestinal Metabolome in the Colon Contents
3.5. FAM Promoted Butyric Acid Production and Enhanced Mucosal Immune Function
3.6. FAM Upregulated IL-22 Production and Enhanced GPR41 and GPR43 Activation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Blikslager, A.T.; Moeser, A.J.; Gookin, J.L.; Jones, S.L.; Odle, J. Restoration of barrier function in injured intestinal mucosa. Physiol. Rev. 2007, 87, 545–564. [Google Scholar] [CrossRef]
- Catassi, C.; Bonucci, A.; Coppa, G.V.; Carlucci, A.; Giorgi, P.L. Intestinal permeability changes during the first month: Effect of natural versus artificial feeding. J. Pediatr. Gastroenterol. Nutr. 1995, 21, 383–386. [Google Scholar] [CrossRef]
- Edelblum, K.L.; Turner, J.R. The tight junction in inflammatory disease: Communication breakdown. Curr. Opin. Pharm. 2009, 9, 715–720. [Google Scholar] [CrossRef] [Green Version]
- Marchiando, A.; Graham, W.V.; Turner, J. Epithelial Barriers in Homeostasis and Disease. Annu. Rev. Pathol. 2010, 5, 119–144. [Google Scholar] [CrossRef]
- Turner, J.R. Intestinal mucosal barrier function in health and disease. Nat. Rev. Immunol. 2009, 9, 799–809. [Google Scholar] [CrossRef]
- Gustafsson, J.K.; Johansson, M.E.V. The role of goblet cells and mucus in intestinal homeostasis. Nat. Rev. Gastro. Hepat. 2022, 1–19. [Google Scholar] [CrossRef]
- Bevins, C.L.; Salzman, N.H. Paneth cells, antimicrobial peptides and maintenance of intestinal homeostasis. Nat. Rev. Microbiol. 2011, 9, 356–368. [Google Scholar] [CrossRef]
- Bonaz, B.L.; Bernstein, C.N. Brain-gut interactions in inflammatory bowel disease. Gastroenterology 2013, 144, 36–49. [Google Scholar] [CrossRef] [Green Version]
- Nicholson, J.K.; Holmes, E.; Kinross, J.; Burcelin, R.; Gibson, G.; Jia, W.; Pettersson, S. Host-gut microbiota metabolic interactions. Science 2012, 336, 1262–1267. [Google Scholar] [CrossRef] [Green Version]
- Maruvada, P.; Leone, V.; Kaplan, L.M.; Chang, E.B. The Human Microbiome and Obesity: Moving beyond Associations. Cell Host Microbe 2017, 22, 589–599. [Google Scholar] [CrossRef]
- Chen, L.; Wang, D.; Garmaeva, S.; Kurilshikov, A.; Vich Vila, A.; Gacesa, R.; Sinha, T.; Segal, E.; Weersma, R.K.; Wijmenga, C.; et al. The long-term genetic stability and individual specificity of the human gut microbiome. Cell 2021, 184, 2302–2315. [Google Scholar] [CrossRef]
- Yano, J.M.; Yu, K.; Donaldson, G.P.; Shastri, G.G.; Ann, P.; Ma, L.; Nagler, C.; Ismagilov, R.; Mazmanian, S.; Hsiao, E.; et al. Indigenous Bacteria from the Gut Microbiota Regulate Host Serotonin Biosynthesis. Cell 2015, 161, 264–276. [Google Scholar] [CrossRef] [Green Version]
- Schirmer, M.; Garner, A.; Vlamakis, H.; Xavier, R.J. Microbial genes and pathways in inflammatory bowel disease. Nat. Rev. Microbiol. 2019, 17, 497–511. [Google Scholar] [CrossRef]
- Willing, B.P.; Dicksved, J.; Halfvarson, J.; Andersson, A.F.; Lucio, M.; Zheng, Z.; Järnerot, G.; Tysk, C.; Jansson, J.K.; Engstrand, L. A pyrosequencing study in twins shows that gastrointestinal microbial profiles vary with inflammatory bowel disease phenotypes. Gastroenterology 2010, 139, 1844–1854. [Google Scholar] [CrossRef]
- Turnbaugh, P.J.; Bäckhed, F.; Fulton, L.; Gordon, J.I. Diet-Induced Obesity Is Linked to Marked but Reversible Alterations in the Mouse Distal Gut Microbiome. Cell Host Microbe 2008, 3, 213–223. [Google Scholar] [CrossRef] [Green Version]
- Carroll, I.M.; Ringel-Kulka, T.; Siddle, J.P.; Ringel, Y. Alterations in composition and diversity of the intestinal microbiota in patients with diarrhea-predominant irritable bowel syndrome. Neurogastroenterol. Motil. 2012, 24, 248–521. [Google Scholar] [CrossRef] [Green Version]
- Mosca, A.; Leclerc, M.; Hugot, J.P. Gut Microbiota Diversity and Human Diseases: Should We Reintroduce Key Predators in Our Ecosystem? Front. Microbiol. 2016, 7, 455. [Google Scholar] [CrossRef] [Green Version]
- Hegarty, J.W.; Guinane, C.M.; Ross, R.P.; Hill, C.; Cotter, P.D. Bacteriocin production: A relatively unharnessed probiotic trait? F1000Res 2016, 5, 2587. [Google Scholar] [CrossRef]
- van Baarlen, P.; Wells, J.M.; Kleerebezem, M. Regulation of intestinal homeostasis and immunity with probiotic lactobacilli. Trends Immunol. 2013, 34, 208–215. [Google Scholar] [CrossRef]
- Klaenhammer, T.R.; Kleerebezem, M.; Kopp, M.V.; Rescigno, M. The impact of probiotics and prebiotics on the immune system. Nat. Rev. Immunol. 2012, 12, 728–734. [Google Scholar] [CrossRef]
- Bali, V.; Panesar, P.S.; Bera, M.B.; Kennedy, J.F. Bacteriocins: Recent Trends and Potential Applications. Crit. Rev. Food Sci. Nutr. 2016, 56, 817–834. [Google Scholar] [CrossRef]
- Rios-Covian, D.; Ruas-Madiedo, P.; Margolles, A.; Gueimonde, M.; de Los, R.C.; Salazar, N. Intestinal Short Chain Fatty Acids and their Link with Diet and Human Health. Front. Microbiol. 2016, 7, 185. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sanna, S.; van Zuydam, N.R.; Mahajan, A.; Kurilshikov, A.; Vich, V.A.; Vosa, U.; Mujagic, Z.; Masclee, A.A.M.; Jonkers, D.M.A.E.; Oosting, M.; et al. Causal relationships among the gut microbiome, short-chain fatty acids and metabolic diseases. Nat. Genet. 2019, 51, 600–605. [Google Scholar] [CrossRef] [PubMed]
- Han, X.; Lee, A.; Huang, S.; Gao, J.; Spence, J.R.; Owyang, C. Lactobacillus rhamnosus GG prevents epithelial barrier dysfunction induced by interferon-gamma and fecal supernatants from irritable bowel syndrome patients in human intestinal enteroids and colonoids. Gut. Microbes 2019, 10, 59–76. [Google Scholar] [CrossRef] [Green Version]
- Roura, E.; Koopmans, S.; Lallès, J.; Le Huerou-Luron, I.; de Jager, N.; Schuurman, T.; Val-Laillet, D. Critical review evaluating the pig as a model for human nutritional physiology. Nutr. Res. Rev. 2016, 29, 60–90. [Google Scholar] [CrossRef]
- Xiao, L.; Estellé, J.; Kiilerich, P.; Ramayo-Caldas, Y.; Xia, Z.; Feng, Q.; Liang, S.; Pedersen, A.O.; Kjeldsen, N.J.; Liu, C.; et al. A reference gene catalogue of the pig gut microbiome. Nat. Microbiol. 2016, 1, 16161. [Google Scholar] [CrossRef] [Green Version]
- Geng, S.; Cheng, S.; Li, Y.; Wen, Z.; Ma, X.; Jiang, X. Faecal Microbiota Transplantation Reduces Susceptibility to Epithelial Injury and Modulates Tryptophan Metabolism of the Microbial Community in a Piglet Model. J. Crohn’s Colitis 2018, 12, 1359–1374. [Google Scholar] [CrossRef]
- Huang, J.; Qi, Y.; Wang, A.; Huang, C.; Liu, X.; Yang, X.; Li, L.; Zhou, R. Porcine β-defensin 2 inhibits proliferation of pseudorabies virus in vitro and in transgenic mice. Virol. J. 2020, 17, 18. [Google Scholar] [CrossRef]
- Casula, G.; Cutting, S.M. Bacillus Probiotics: Spore Germination in the Gastrointestinal Tract. Appl. Environ. Microb. 2002, 68, 2344–2352. [Google Scholar] [CrossRef] [Green Version]
- Sivieri, K.; Morales, M.L.V.; Adorno, M.A.T.; Sakamoto, I.K.; Saad, S.M.I.; Rossi, E.A. Lactobacillus acidophilus CRL 1014 improved “gut health” in the SHIME® reactor. BMC Gastroenterol. 2013, 13, 100. [Google Scholar] [CrossRef]
- Luise, D.; Bertocchi, M.; Motta, V.; Salvarani, C.; Bosi, P.; Luppi, A.; Fanelli, F.; Mazzoni, M.; Archetti, I.; Maiorano, G.; et al. Bacillus sp. probiotic supplementation diminish the Escherichia coli F4ac infection in susceptible weaned pigs by influencing the intestinal immune response, intestinal microbiota and blood metabolomics. J. Anim. Sci. Biotechnol. 2019, 10, 74. [Google Scholar] [CrossRef] [PubMed]
- Salinas, I.; Cuesta, A.; Esteban, M.Á.; Meseguer, J. Dietary administration of Lactobacillus delbrüeckii and Bacillus subtilis, single or combined, on gilthead seabream cellular innate immune responses. Fish Shellfish Immun. 2005, 19, 67–77. [Google Scholar] [CrossRef]
- Røssland, E.; Langsrud, T.; Granum, P.E.; Sørhaug, T. Production of antimicrobial metabolites by strains of Lactobacillus or Lactococcus co-cultured with Bacillus cereus in milk. Int. J. Food Microbiol. 2005, 98, 193–200. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, C.; You, F.; Dai, F.; Tung, Y.; Chen, W.; Chu, H.; Wu, S.-H.; Vhau, C.-F. Bacillus coagulans BC198 and Lactobacillus paracasei S38 in combination reduce body fat accumulation and modulate gut microbiota. CyTA-J. Food 2020, 18, 764–775. [Google Scholar] [CrossRef]
- Severson, E.A.; Kwon, M.; Hilgarth, R.S.; Parkos, C.A.; Nusrat, A. Glycogen Synthase Kinase 3 (GSK-3) influences epithelial barrier function by regulating Occludin, Claudin-1 and E-cadherin expression. Biochem. Biophys. Res. Commun. 2010, 397, 592–597. [Google Scholar] [CrossRef] [Green Version]
- Gumbiner, B. Structure, biochemistry, and assembly of epithelial tight junctions. Am. J. Physiol.-Cell Physiol. 1987, 253, C749–C758. [Google Scholar] [CrossRef] [PubMed]
- Schmitz, H.; Barmeyer, C.; Fromm, M.; Runkel, N.; Foss, H.; Bentzel, C.J.; Riechen, E.-O.; Schulzke, J.-D. Altered tight junction structure contributes to the impaired epithelial barrier function in ulcerative colitis. Gastroenterology 1999, 116, 301–309. [Google Scholar] [CrossRef]
- McDole, J.R.; Wheeler, L.W.; McDonald, K.G.; Wang, B.; Konjufca, V.; Knoop, K.A. Goblet cells deliver luminal antigen to CD103+ dendritic cells in the small intestine. Nature 2012, 483, 345–349. [Google Scholar] [CrossRef] [Green Version]
- Cash, H.L.; Whitham, C.V.; Behrendt, C.L.; Hooper, L.V. Symbiotic Bacteria Direct Expression of an Intestinal Bactericidal Lectin. Science 2006, 313, 1126–1130. [Google Scholar] [CrossRef] [Green Version]
- Fan, L.; Qi, Y.; Qu, S.; Chen, X.; Li, A.; Hendi, M.; Xu, C.; Wang, L.; Hou, T.; Si, J.; et al. B. adolescentis ameliorates chronic colitis by regulating Treg/Th2 response and gut microbiota remodeling. Gut Microbes 2021, 13, 1–17. [Google Scholar] [CrossRef]
- Molist, F.; Manzanilla, E.G.; Pérez, J.F.; Nyachoti, C.M. Coarse, but not finely ground, dietary fibre increases intestinal Firmicutes: Bacteroidetes ratio and reduces diarrhoea induced by experimental infection in piglets. Br. J. Nutr. 2012, 108, 9–15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pickard, J.M.; Zeng, M.Y.; Caruso, R.; Nunez, G. Gut microbiota: Role in pathogen colonization, immune responses, and inflammatory disease. Immunol. Rev. 2017, 279, 70–89. [Google Scholar] [CrossRef] [PubMed]
- Shapiro, H.; Kolodziejczyk, A.A.; Halstuch, D.; Elinav, E. Bile acids in glucose metabolism in health and disease. J. Exp. Med. 2018, 215, 383–396. [Google Scholar] [CrossRef] [PubMed]
- Lagkouvardos, I.; Lesker, T.R.; Hitch, T.; Galvez, E.; Smit, N.; Neuhaus, K.; Wang, J.; Baines, J.F.; Abt, B.; Stecher, B.; et al. Sequence and cultivation study of Muribaculaceae reveals novel species, host preference, and functional potential of this yet undescribed family. Microbiome 2019, 7, 28. [Google Scholar] [CrossRef] [Green Version]
- Andersen, V.; Olsen, A.; Carbonnel, F.; Tjønneland, A.; Vogel, U. Diet and risk of inflammatory bowel disease. Dig. Liver Dis. 2012, 44, 185–194. [Google Scholar] [CrossRef]
- Ye, J.; Lv, L.; Wu, W.; Li, Y.; Shi, D.; Fang, D.; Guo, F.; Jiang, H.; Yan, R.; Ye, W.; et al. Butyrate protects mice against methionine–choline-deficient diet-induced non-alcoholic steatohepatitis by improving gut barrier function, attenuating inflammation and reducing endotoxin levels. Front. Microbiol. 2018, 9, 1967. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.; Zhang, L.; Ye, X.; Chen, L.; Zhang, L.; Gao, Y.; Kang, J.X.; Cai, C. Characteristics of fatty acid distribution is associated with colorectal cancer prognosis. Prostaglandins Leukot. Essent. Fatty Acid. 2013, 88, 355–360. [Google Scholar] [CrossRef]
- Kim, H.K.; Rutten, N.; Besseling-van Der Vaart, I.; Niers, L.; Choi, Y.H.; Rijkers, G.T.; van Hemert, S. Probiotic supplementation influences faecal short chain fatty acids in infants at high risk for eczema. Benef. Microbes 2015, 6, 783–790. [Google Scholar] [CrossRef]
- Markowiak-Kopeć, P.; Śliżewska, K. The effect of probiotics on the production of short-chain fatty acids by human intestinal microbiome. Nutrients 2020, 12, 1107. [Google Scholar] [CrossRef] [Green Version]
- Zhuang, M.; Shang, W.; Ma, Q.; Strappe, P.; Zhou, Z. Abundance of probiotics and butyrate-production microbiome manages constipation via short-chain fatty acids production and hormones secretion. Mol. Nutr. Food Res. 2019, 63, 1801187. [Google Scholar] [CrossRef]
- Ratajczak, W.; Rył, A.; Mizerski, A.; Walczakiewicz, K.; Sipak, O.; Laszczyńska, M. Immunomodulatory potential of gut microbiome-derived short-chain fatty acids (SCFAs). Acta Biochim. Polon. 2019, 66, 1–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, L.; Sun, M.; Wu, W.; Yang, W.; Huang, X.; Xiao, Y.; Ma, C.; Xu, L.; Yao, S.; Liu, Z.; et al. Microbiota metabolite butyrate differentially regulates Th1 and Th17 cells’ differentiation and function in induction of colitis. Inflamm. Bowel Dis. 2019, 25, 1450–1461. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smith, P.M.; Howitt, M.R.; Panikov, N.; Michaud, M.; Gallini, C.A.; Bohlooly-Y, M.; Glikman, J.N.; Garrett, W.S. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science 2013, 341, 569–573. [Google Scholar] [CrossRef] [Green Version]
- Sun, M.; Wu, W.; Chen, L.; Yang, W.; Huang, X.; Ma, C.; Chen, F.; Xiao, Y.; Zhao, Y.; Ma, C.; et al. Microbiota-derived short-chain fatty acids promote Th1 cell IL-10 production to maintain intestinal homeostasis. Nat. Commun. 2018, 9, 3555. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Min, Y.N.; Yang, H.L.; Xu, Y.X.; Gao, Y.P. Effects of dietary supplementation of synbiotics on growth performance, intestinal morphology, sIgA content and antioxidant capacities of broilers. J. Anim. Physiol. Anim. Nutr. 2016, 100, 1073–1080. [Google Scholar] [CrossRef]
- Xiao, L.; Gong, C.; Ding, Y.; Ding, G.; Xu, X.; Deng, C.; Ze, X.; Malard, P.; Ben, X. Probiotics maintain intestinal secretory immunoglobulin A levels in healthy formula-fed infants: A randomised, double-blind, placebo-controlled study. Benef. Microbes 2019, 10, 729–739. [Google Scholar] [CrossRef]
- Sun, M.; Wu, W.; Liu, Z.; Cong, Y. Microbiota metabolite short chain fatty acids, GPCR., and inflammatory bowel diseases. J. Gastroenterol. 2017, 52, 1–8. [Google Scholar] [CrossRef]
- Zhao, Y.; Chen, F.; Wu, W.; Sun, M.; Bilotta, A.J.; Yao, S.; Xiao, Y.; Huang, X.; Eaves-Pyles, T.D.; Golovko, G.; et al. GPR43 mediates microbiota metabolite SCFA regulation of antimicrobial peptide expression in intestinal epithelial cells via activation of mTOR and STAT3. Mucosal Immunol. 2018, 11, 752–762. [Google Scholar] [CrossRef] [Green Version]
- Rooks, M.G.; Garrett, W.S. Gut microbiota, metabolites and host immunity. Nat. Rev. Immunol. 2016, 16, 341–352. [Google Scholar] [CrossRef]







| Item | Control | FAM | Antibiotic |
|---|---|---|---|
| Initial BW, kg | 10.34 ± 0.45 | 10.02 ± 0.46 | 9.96 ± 0.51 |
| Final BW, kg | 23.75 ± 2.99 | 25.98 ± 3.31 | 24.71 ± 2.85 |
| ADG, g/d | 447 ± 22.3 b | 532 ± 28.5 a | 492 ± 19.7 a |
| ADFI, g/d | 688 ± 45.7 ab | 736 ± 47.8 a | 686 ± 38.6 ab |
| F/G | 1.53 ± 0.05 a | 1.36 ± 0.02 b | 1.39 ± 0.03 b |
| Diarrhea incidence | 8.00 ± 0.50 a | 4.14 ± 0.25 b | 4.47 ± 0.32 b |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xie, Z.; Li, M.; Qian, M.; Yang, Z.; Han, X. Co-Cultures of Lactobacillus acidophilus and Bacillus subtilis Enhance Mucosal Barrier by Modulating Gut Microbiota-Derived Short-Chain Fatty Acids. Nutrients 2022, 14, 4475. https://doi.org/10.3390/nu14214475
Xie Z, Li M, Qian M, Yang Z, Han X. Co-Cultures of Lactobacillus acidophilus and Bacillus subtilis Enhance Mucosal Barrier by Modulating Gut Microbiota-Derived Short-Chain Fatty Acids. Nutrients. 2022; 14(21):4475. https://doi.org/10.3390/nu14214475
Chicago/Turabian StyleXie, Zhengjun, Meng Li, Mengqi Qian, Zhiren Yang, and Xinyan Han. 2022. "Co-Cultures of Lactobacillus acidophilus and Bacillus subtilis Enhance Mucosal Barrier by Modulating Gut Microbiota-Derived Short-Chain Fatty Acids" Nutrients 14, no. 21: 4475. https://doi.org/10.3390/nu14214475
APA StyleXie, Z., Li, M., Qian, M., Yang, Z., & Han, X. (2022). Co-Cultures of Lactobacillus acidophilus and Bacillus subtilis Enhance Mucosal Barrier by Modulating Gut Microbiota-Derived Short-Chain Fatty Acids. Nutrients, 14(21), 4475. https://doi.org/10.3390/nu14214475