Cranberry Polyphenols in Esophageal Cancer Inhibition: New Insights
Abstract
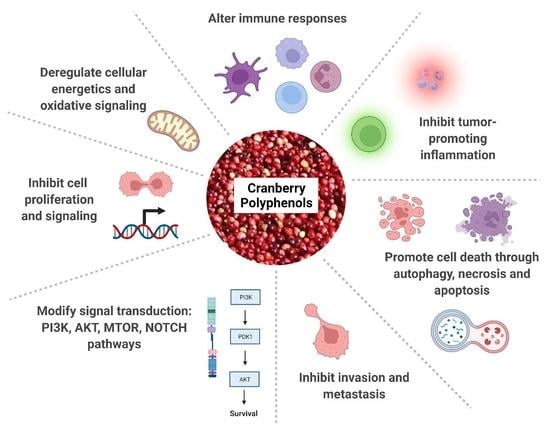
:1. Introduction
2. Materials and Methods
2.1. Cranberry Polyphenol Preparation
2.2. Cell Lines and Dose Determination Using Calcein-AM Based Viability Assay
2.3. Lysate Collection and Western Blot Analysis of BE and EAC Cells Treated with Cranberry Polyphenols
2.4. Functional Proteomics by RPPA
2.5. Bioinformatic Analysis of RPPA Data
2.6. Statistical Analyses
3. Results
3.1. Cranberry Polyphenols Inhibit Cell Viability of Premalignant Barrett’s Esophageal and Adenocarcinoma Derived Cell Lines
3.2. RPPA Analysis of Lysates from Esophageal Adenocarcinoma Cells Treated with C-PAC or AFG Reveal Modulation of Numerous Cancer-Linked Processes
3.3. Cranberry Polyphenols Modulate P53-Linked, Inflammatory and Cell Cycle Proteins in BE and EAC Cells
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ustaoglu, A.; Nguyen, A.; Spechler, S.; Sifrim, D.; Souza, R.; Woodland, P. Mucosal pathogenesis in gastro-esophageal reflux disease. Neurogastroenterol. Motil. 2020, 32, e14022. [Google Scholar] [CrossRef] [PubMed]
- Napier, K.J.; Scheerer, M.; Misra, S. Esophageal cancer: A Review of epidemiology, pathogenesis, staging workup and treatment modalities. World J. Gastrointest. Oncol. 2014, 6, 112–120. [Google Scholar] [CrossRef] [PubMed]
- Dulak, A.M.; Stojanov, P.; Peng, S.; Lawrence, M.S.; Fox, C.; Stewart, C.; Bandla, S.; Imamura, Y.; Schumacher, S.E.; Shefler, E.; et al. Exome and whole-genome sequencing of esophageal adenocarcinoma identifies recurrent driver events and mutational complexity. Nat. Genet. 2013, 45, 478–486. [Google Scholar] [CrossRef] [PubMed]
- Grady, W.M.; Yu, M.; Markowitz, S.D.; Chak, A. Barrett’s Esophagus and Esophageal Adenocarcinoma Biomarkers. Cancer Epidemiol. Biomark. Prev. 2020, 29, 2486–2494. [Google Scholar] [CrossRef]
- Contino, G.; Vaughan, T.L.; Whiteman, D.; Fitzgerald, R.C. The Evolving Genomic Landscape of Barrett’s Esophagus and Esophageal Adenocarcinoma. Gastroenterology 2017, 153, 657–673.e651. [Google Scholar] [CrossRef]
- Domper Arnal, M.J.; Ferrandez Arenas, A.; Lanas Arbeloa, A. Esophageal cancer: Risk factors, screening and endoscopic treatment in Western and Eastern countries. World J. Gastroenterol. 2015, 21, 7933–7943. [Google Scholar] [CrossRef]
- American Cancer Society. Cancer Facts & Figures 2021; American Cancer Society: Atlanta, GA, USA, 2021. [Google Scholar]
- Howell, A.B.; Reed, J.D.; Krueger, C.G.; Winterbottom, R.; Cunningham, D.G.; Leahy, M. A-type cranberry proanthocyanidins and uropathogenic bacterial anti-adhesion activity. Phytochemistry 2005, 66, 2281–2291. [Google Scholar] [CrossRef]
- Greenberg, J.A.; Newmann, S.J.; Howell, A.B. Consumption of sweetened dried cranberries versus unsweetened raisins for inhibition of uropathogenic Escherichia coli adhesion in human urine: A pilot study. J. Altern. Complement. Med. 2005, 11, 875–878. [Google Scholar] [CrossRef]
- Di Martino, P.; Agniel, R.; David, K.; Templer, C.; Gaillard, J.L.; Denys, P.; Botto, H. Reduction of Escherichia coli adherence to uroepithelial bladder cells after consumption of cranberry juice: A double-blind randomized placebo-controlled cross-over trial. World J. Urol. 2006, 24, 21–27. [Google Scholar] [CrossRef]
- Weh, K.M.; Clarke, J.; Kresty, L.A. Cranberries and Cancer: An Update of Preclinical Studies Evaluating the Cancer Inhibitory Potential of Cranberry and Cranberry Derived Constituents. Antioxidants 2016, 5, 27. [Google Scholar] [CrossRef]
- Jozkowiak, M.; Skupin-Mrugalska, P.; Nowicki, A.; Borys-Wojcik, S.; Wierzchowski, M.; Kaczmarek, M.; Ramlau, P.; Jodynis-Liebert, J.; Piotrowska-Kempisty, H. The Effect of 4-hydroxy-3,4,5-trimetoxystilbene, the Metabolite of Resveratrol Analogue DMU-212, on Growth, Cell Cycle and Apoptosis in DLD-1 and LOVO Colon Cancer Cell Lines. Nutrients 2020, 12, 1327. [Google Scholar] [CrossRef]
- Mansouri, R.A.; Percival, S.S. Cranberry extract initiates intrinsic apoptosis in HL-60 cells by increasing BAD activity through inhibition of AKT phosphorylation. BMC Complement. Med. Ther. 2020, 20, 71. [Google Scholar] [CrossRef] [Green Version]
- Khairnar, M.R.; Wadgave, U.; Jadhav, H.; Naik, R. Anticancer activity of chlorhexidine and cranberry extract: An in-vitro study. J. Exp. Ther. Oncol. 2018, 12, 201–205. [Google Scholar]
- Prasain, J.K.; Rajbhandari, R.; Keeton, A.B.; Piazza, G.A.; Barnes, S. Metabolism and growth inhibitory activity of cranberry derived flavonoids in bladder cancer cells. Food Funct. 2016, 7, 4012–4019. [Google Scholar] [CrossRef] [Green Version]
- Wu, X.; Song, M.; Cai, X.; Neto, C.; Tata, A.; Han, Y.; Wang, Q.; Tang, Z.; Xiao, H. Chemopreventive Effects of Whole Cranberry (Vaccinium macrocarpon) on Colitis-Associated Colon Tumorigenesis. Mol. Nutr. Food Res. 2018, 62, e1800942. [Google Scholar] [CrossRef]
- Wu, X.; Xue, L.; Tata, A.; Song, M.; Neto, C.C.; Xiao, H. Bioactive Components of Polyphenol-Rich and Non-Polyphenol-Rich Cranberry Fruit Extracts and Their Chemopreventive Effects on Colitis-Associated Colon Cancer. J. Agric. Food Chem. 2020, 68, 6845–6853. [Google Scholar] [CrossRef]
- Jin, D.; Liu, T.; Dong, W.; Zhang, Y.; Wang, S.; Xie, R.; Wang, B.; Cao, H. Dietary feeding of freeze-dried whole cranberry inhibits intestinal tumor development in Apcmin/+ mice. Oncotarget 2017, 8, 97787–97800. [Google Scholar] [CrossRef] [Green Version]
- Kresty, L.A.; Clarke, J.; Ezell, K.; Exum, A.; Howell, A.B.; Guettouche, T. MicroRNA alterations in Barrett’s esophagus, esophageal adenocarcinoma, and esophageal adenocarcinoma cell lines following cranberry extract treatment: Insights for chemoprevention. J. Carcinog. 2011, 10, 34. [Google Scholar] [CrossRef]
- Kresty, L.A.; Weh, K.M.; Zeyzus-Johns, B.; Perez, L.N.; Howell, A.B. Cranberry proanthocyanidins inhibit esophageal adenocarcinoma in vitro and in vivo through pleiotropic cell death induction and PI3K/AKT/mTOR inactivation. Oncotarget 2015, 6, 33438–33455. [Google Scholar] [CrossRef] [Green Version]
- Stevenson, D.E.; Hurst, R.D. Polyphenolic phytochemicals—Just antioxidants or much more? Cell Mol. Life Sci. 2007, 64, 2900–2916. [Google Scholar] [CrossRef]
- Weh, K.M.; Aiyer, H.S.; Howell, A.B.; Kresty, L.A. Cranberry proanthocyanidins modulate reactive oxygen species in Barrett’s and esophageal adenocarcinoma cell lines. J. Berry Res. 2016, 6, 125–136. [Google Scholar] [CrossRef] [Green Version]
- Foo, L.Y.; Lu, Y.; Howell, A.B.; Vorsa, N. A-Type proanthocyanidin trimers from cranberry that inhibit adherence of uropathogenic P-fimbriated Escherichia coli. J. Nat. Prod. 2000, 63, 1225–1228. [Google Scholar] [CrossRef]
- Krueger, C.G.; Reed, J.D.; Feliciano, R.P.; Howell, A.B. Quantifying and characterizing proanthocyanidins in cranberries in relation to urinary tract health. Anal. Bioanal. Chem. 2013, 405, 4385–4395. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Howell, A.B.; Zhang, D.J.; Khoo, C. A randomized, double-blind, placebo-controlled pilot study to assess bacterial anti-adhesive activity in human urine following consumption of a cranberry supplement. Food Funct. 2019, 10, 7645–7652. [Google Scholar] [CrossRef] [Green Version]
- Brown, P.N.; Shipley, P.R. Determination of anthocyanins in cranberry fruit and cranberry fruit products by high-performance liquid chromatography with ultraviolet detection: Single-laboratory validation. J. AOAC Int. 2011, 94, 459–466. [Google Scholar] [CrossRef] [Green Version]
- Suchorolski, M.T.; Paulson, T.G.; Sanchez, C.A.; Hockenbery, D.; Reid, B.J. Warburg and Crabtree effects in premalignant Barrett’s esophagus cell lines with active mitochondria. PLoS ONE 2013, 8, e56884. [Google Scholar] [CrossRef] [Green Version]
- Alvarez, H.; Koorstra, J.B.; Hong, S.M.; Boonstra, J.J.; Dinjens, W.N.; Foratiere, A.A.; Wu, T.T.; Montgomery, E.; Eshleman, J.R.; Maitra, A. Establishment and characterization of a bona fide Barrett esophagus-associated adenocarcinoma cell line. Cancer Biol. Ther. 2008, 7, 1753–1755. [Google Scholar]
- Weh, K.M.; Howell, A.B.; Kresty, L.A. Expression, modulation, and clinical correlates of the autophagy protein Beclin-1 in esophageal adenocarcinoma. Mol. Carcinog. 2016, 55, 1876–1885. [Google Scholar] [CrossRef]
- Barbhuiya, M.A.; Kashyap, M.K.; Puttamallesh, V.N.; Kumar, R.V.; Wu, X.; Pandey, A.; Gowda, H. Identification of spleen tyrosine kinase as a potential therapeutic target for esophageal squamous cell carcinoma using reverse phase protein arrays. Oncotarget 2018, 9, 18422–18434. [Google Scholar] [CrossRef] [Green Version]
- Paweletz, C.P.; Charboneau, L.; Bichsel, V.E.; Simone, N.L.; Chen, T.; Gillespie, J.W.; Emmert-Buck, M.R.; Roth, M.J.; Petricoin, I.E.; Liotta, L.A. Reverse phase protein microarrays which capture disease progression show activation of pro-survival pathways at the cancer invasion front. Oncogene 2001, 20, 1981–1989. [Google Scholar] [CrossRef] [Green Version]
- Fouad, Y.A.; Aanei, C. Revisiting the hallmarks of cancer. Am. J. Cancer Res. 2017, 7, 1016–1036. [Google Scholar]
- Fichter, C.D.; Herz, C.; Munch, C.; Opitz, O.G.; Werner, M.; Lassmann, S. Occurrence of multipolar mitoses and association with Aurora-A/-B kinases and p53 mutations in aneuploid esophageal carcinoma cells. BMC Cell Biol. 2011, 12, 13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bensaad, K.; Tsuruta, A.; Selak, M.A.; Vidal, M.N.; Nakano, K.; Bartrons, R.; Gottlieb, E.; Vousden, K.H. TIGAR, a p53-inducible regulator of glycolysis and apoptosis. Cell 2006, 126, 107–120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, H.P.; Xie, J.M.; Li, B.; Sun, Y.H.; Gao, Q.G.; Ding, Z.H.; Wu, H.R.; Qin, Z.H. TIGAR regulates DNA damage and repair through pentosephosphate pathway and Cdk5-ATM pathway. Sci. Rep. 2015, 5, 9853. [Google Scholar] [CrossRef] [Green Version]
- Mileo, A.M.; Miccadei, S. Polyphenols as Modulator of Oxidative Stress in Cancer Disease: New Therapeutic Strategies. Oxid. Med. Cell Longev. 2016, 2016, 6475624. [Google Scholar] [CrossRef] [Green Version]
- Xu, Y.; Surman, D.R.; Diggs, L.; Xi, S.; Gao, S.; Gurusamy, D.; McLoughlin, K.; Drake, J.; Feingold, P.; Brown, K.; et al. Bile acid-induced “Minority MOMP” promotes esophageal carcinogenesis while maintaining apoptotic resistance via Mcl-1. Oncogene 2020, 39, 877–890. [Google Scholar] [CrossRef]
- Fang, X.; Yu, S.; Eder, A.; Mao, M.; Bast, R.C., Jr.; Boyd, D.; Mills, G.B. Regulation of BAD phosphorylation at serine 112 by the Ras-mitogen-activated protein kinase pathway. Oncogene 1999, 18, 6635–6640. [Google Scholar] [CrossRef] [Green Version]
- Dasika, G.K.; Lin, S.C.; Zhao, S.; Sung, P.; Tomkinson, A.; Lee, E.Y. DNA damage-induced cell cycle checkpoints and DNA strand break repair in development and tumorigenesis. Oncogene 1999, 18, 7883–7899. [Google Scholar] [CrossRef] [Green Version]
- Peterson, R.T.; Schreiber, S.L. Translation control: Connecting mitogens and the ribosome. Curr. Biol. 1998, 8, 248–250. [Google Scholar] [CrossRef] [Green Version]
- Dufner, A.; Thomas, G. Ribosomal S6 kinase signaling and the control of translation. Exp. Cell Res. 1999, 253, 100–109. [Google Scholar] [CrossRef]
- Meyuhas, O. Ribosomal Protein S6 Phosphorylation: Four Decades of Research. Int. Rev. Cell Mol. Biol. 2015, 320, 41–73. [Google Scholar] [PubMed]
- Meurette, O.; Stylianou, S.; Rock, R.; Collu, G.M.; Gilmore, A.P.; Brennan, K. Notch activation induces Akt signaling via an autocrine loop to prevent apoptosis in breast epithelial cells. Cancer Res. 2009, 69, 5015–5022. [Google Scholar] [CrossRef] [Green Version]
- Gutierrez, A.; Look, A.T. NOTCH and PI3K-AKT pathways intertwined. Cancer Cell 2007, 12, 411–413. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Straub, T.J.; Chou, W.C.; Manson, A.L.; Schreiber, H.L.; Walker, B.J.; Desjardins, C.A.; Chapman, S.B.; Kaspar, K.L.; Kahsai, O.J.; Traylor, E.; et al. Limited effects of long-term daily cranberry consumption on the gut microbiome in a placebo-controlled study of women with recurrent urinary tract infections. BMC Microbiol. 2021, 21, 53. [Google Scholar] [CrossRef]
- Li, Z.X.; Ma, J.L.; Guo, Y.; Liu, W.D.; Li, M.; Zhang, L.F.; Zhang, Y.; Zhou, T.; Zhang, J.Y.; Gao, H.E.; et al. Suppression of Helicobacter pylori infection by daily cranberry intake: A double-blind, randomized, placebo-controlled trial. J. Gastroenterol. Hepatol. 2021, 36, 927–935. [Google Scholar] [CrossRef]
- Maki, K.C.; Kaspar, K.L.; Khoo, C.; Derrig, L.H.; Schild, A.L.; Gupta, K. Consumption of a cranberry juice beverage lowered the number of clinical urinary tract infection episodes in women with a recent history of urinary tract infection. Am. J. Clin. Nutr. 2016, 103, 1434–1442. [Google Scholar] [CrossRef] [Green Version]
- Tao, W.; Zhang, Y.; Shen, X.; Cao, Y.; Shi, J.; Ye, X.; Chen, S. Rethinking the Mechanism of the Health Benefits of Proanthocyanidins: Absorption, Metabolism, and Interaction with Gut Microbiota. Compr. Rev. Food Sci. Food Saf. 2019, 18, 971–985. [Google Scholar] [CrossRef] [Green Version]
- Bekiares, N.; Krueger, C.G.; Meudt, J.J.; Shanmuganayagam, D.; Reed, J.D. Effect of Sweetened Dried Cranberry Consumption on Urinary Proteome and Fecal Microbiome in Healthy Human Subjects. OMICS 2018, 22, 145–153. [Google Scholar] [CrossRef] [Green Version]
- Student, V.; Vidlar, A.; Bouchal, J.; Vrbkova, J.; Kolar, Z.; Kral, M.; Kosina, P.; Vostalova, J. Cranberry intervention in patients with prostate cancer prior to radical prostatectomy. Clinical, pathological and laboratory findings. Biomed. Pap. Med. Fac. Univ. Palacky Olomouc. Czech Repub. 2016, 160, 559–565. [Google Scholar] [CrossRef] [Green Version]
- Ferguson, P.J.; Kurowska, E.M.; Freeman, D.J.; Chambers, A.F.; Koropatnick, J. In vivo inhibition of growth of human tumor lines by flavonoid fractions from cranberry extract. Nutr. Cancer 2006, 56, 86–94. [Google Scholar] [CrossRef]
- Ferguson, P.J.; Kurowska, E.; Freeman, D.J.; Chambers, A.F.; Koropatnick, D.J. A flavonoid fraction from cranberry extract inhibits proliferation of human tumor cell lines. J. Nutr. 2004, 134, 1529–1535. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, X.; Liu, R.H. Cranberry phytochemicals: Isolation, structure elucidation, and their antiproliferative and antioxidant activities. J. Agric. Food Chem. 2006, 54, 7069–7074. [Google Scholar] [CrossRef] [PubMed]
- Vu, K.D.; Carlettini, H.; Bouvet, J.; Cote, J.; Doyon, G.; Sylvain, J.F.; Lacroix, M. Effect of different cranberry extracts and juices during cranberry juice processing on the antiproliferative activity against two colon cancer cell lines. Food Chem. 2012, 132, 959–967. [Google Scholar] [CrossRef]
- Blumberg, J.B.; Camesano, T.A.; Cassidy, A.; Kris-Etherton, P.; Howell, A.; Manach, C.; Ostertag, L.M.; Sies, H.; Skulas-Ray, A.; Vita, J.A. Cranberries and their bioactive constituents in human health. Adv. Nutr. 2013, 4, 618–632. [Google Scholar] [CrossRef] [Green Version]
- Mungamuri, S.K.; Yang, X.; Thor, A.D.; Somasundaram, K. Survival signaling by Notch1: Mammalian target of rapamycin (mTOR)-dependent inhibition of p53. Cancer Res. 2006, 66, 4715–4724. [Google Scholar] [CrossRef] [Green Version]
- Cecchinato, V.; Chiaramonte, R.; Nizzardo, M.; Cristofaro, B.; Basile, A.; Sherbet, G.V.; Comi, P. Resveratrol-induced apoptosis in human T-cell acute lymphoblastic leukaemia MOLT-4 cells. Biochem. Pharmacol. 2007, 74, 1568–1574. [Google Scholar] [CrossRef]
- Kawahara, T.; Kawaguchi-Ihara, N.; Okuhashi, Y.; Itoh, M.; Nara, N.; Tohda, S. Cyclopamine and quercetin suppress the growth of leukemia and lymphoma cells. Anticancer Res. 2009, 29, 4629–4632. [Google Scholar]
- Pinchot, S.N.; Jaskula-Sztul, R.; Ning, L.; Peters, N.R.; Cook, M.R.; Kunnimalaiyaan, M.; Chen, H. Identification and validation of Notch pathway activating compounds through a novel high-throughput screening method. Cancer 2011, 117, 1386–1398. [Google Scholar] [CrossRef] [Green Version]
- LaFoya, B.; Munroe, J.A.; Albig, A.R. A comparison of resveratrol and other polyphenolic compounds on Notch activation and endothelial cell activity. PLoS ONE 2019, 14, e0210607. [Google Scholar] [CrossRef]
- Yu, X.M.; Jaskula-Sztul, R.; Ahmed, K.; Harrison, A.D.; Kunnimalaiyaan, M.; Chen, H. Resveratrol induces differentiation markers expression in anaplastic thyroid carcinoma via activation of Notch1 signaling and suppresses cell growth. Mol. Cancer Ther. 2013, 12, 1276–1287. [Google Scholar] [CrossRef] [Green Version]
- Wang, Z.; Chen, J.; Capobianco, A.J. The Notch signaling pathway in esophageal adenocarcinoma. Cell Mol. Biol. 2015, 61, 24–32. [Google Scholar] [PubMed]
- Kunze, B.; Wein, F.; Fang, H.Y.; Anand, A.; Baumeister, T.; Strangmann, J.; Gerland, S.; Ingermann, J.; Munch, N.S.; Wiethaler, M.; et al. Notch Signaling Mediates Differentiation in Barrett’s Esophagus and Promotes Progression to Adenocarcinoma. Gastroenterology 2020, 159, 575–590. [Google Scholar] [CrossRef]
- Tamagawa, Y.; Ishimura, N.; Uno, G.; Yuki, T.; Kazumori, H.; Ishihara, S.; Amano, Y.; Kinoshita, Y. Notch signaling pathway and Cdx2 expression in the development of Barrett’s esophagus. Lab. Investig. 2012, 92, 896–909. [Google Scholar] [CrossRef] [PubMed]
- Kunze, B.; Middelhoff, M.; Maurer, H.C.; Agibalova, T.; Anand, A.; Buhrer, A.M.; Fang, H.Y.; Baumeister, T.; Steiger, K.; Strangmann, J.; et al. Notch signaling drives development of Barrett’s metaplasia from Dclk1-positive epithelial tuft cells in the murine gastric mucosa. Sci. Rep. 2021, 11, 4509. [Google Scholar] [CrossRef]
- Wang, Z.; Da Silva, T.G.; Jin, K.; Han, X.; Ranganathan, P.; Zhu, X.; Sanchez-Mejias, A.; Bai, F.; Li, B.; Fei, D.L.; et al. Notch signaling drives stemness and tumorigenicity of esophageal adenocarcinoma. Cancer Res. 2014, 74, 6364–6374. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Monahan, P.; Rybak, S.; Raetzman, L.T. The notch target gene HES1 regulates cell cycle inhibitor expression in the developing pituitary. Endocrinology 2009, 150, 4386–4394. [Google Scholar] [CrossRef] [Green Version]
- Bhat, A.A.; Nisar, S.; Maacha, S.; Carneiro-Lobo, T.C.; Akhtar, S.; Siveen, K.S.; Wani, N.A.; Rizwan, A.; Bagga, P.; Singh, M.; et al. Cytokine-chemokine network driven metastasis in esophageal cancer; promising avenue for targeted therapy. Mol. Cancer 2021, 20, 2. [Google Scholar] [CrossRef]
- Lagisetty, K.H.; McEwen, D.P.; Nancarrow, D.J.; Schiebel, J.G.; Ferrer-Torres, D.; Ray, D.; Frankel, T.L.; Lin, J.; Chang, A.C.; Kresty, L.A.; et al. Immune determinants of Barrett’s progression to esophageal adenocarcinoma. JCI Insight 2021, 6, e143888. [Google Scholar] [CrossRef]
- Milano, F.; Jorritsma, T.; Rygiel, A.M.; Bergman, J.J.; Sondermeijer, C.; Ten Brinke, A.; vanHam, S.M.; Krishnadath, K.K. Expression pattern of immune suppressive cytokines and growth factors in oesophageal adenocarcinoma reveal a tumour immune escape-promoting microenvironment. Scand. J. Immunol. 2008, 68, 616–623. [Google Scholar] [CrossRef]
- Blum, A.E.; Venkitachalam, S.; Ravillah, D.; Chelluboyina, A.K.; Kieber-Emmons, A.M.; Ravi, L.; Kresak, A.; Chandar, A.K.; Markowitz, S.D.; Canto, M.I.; et al. Systems Biology Analyses Show Hyperactivation of Transforming Growth Factor-beta and JNK Signaling Pathways in Esophageal Cancer. Gastroenterology 2019, 156, 1761–1774. [Google Scholar] [CrossRef]
- Ebbing, E.A.; van der Zalm, A.P.; Steins, A.; Creemers, A.; Hermsen, S.; Rentenaar, R.; Klein, M.; Waasdorp, C.; Hooijer, G.K.J.; Meijer, S.L.; et al. Stromal-derived interleukin 6 drives epithelial-to-mesenchymal transition and therapy resistance in esophageal adenocarcinoma. Proc. Natl. Acad. Sci. USA 2019, 116, 2237–2242. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Caldas, A.P.S.; Coelho, O.G.L.; Bressan, J. Cranberry antioxidant power on oxidative stress, inflammation and mitochondrial damage. Int. J. Food Prop. 2018, 21, 582–592. [Google Scholar] [CrossRef]
- Frankell, A.M.; Jammula, S.; Li, X.; Contino, G.; Killcoyne, S.; Abbas, S.; Perner, J.; Bower, L.; Devonshire, G.; Ococks, E.; et al. The landscape of selection in 551 esophageal adenocarcinomas defines genomic biomarkers for the clinic. Nat. Genet. 2019, 51, 506–516. [Google Scholar] [CrossRef] [PubMed]
- Stachler, M.D.; Camarda, N.D.; Deitrick, C.; Kim, A.; Agoston, A.T.; Odze, R.D.; Hornick, J.L.; Nag, A.; Thorner, A.R.; Ducar, M.; et al. Detection of Mutations in Barrett’s Esophagus before Progression to High-Grade Dysplasia or Adenocarcinoma. Gastroenterology 2018, 155, 156–167. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ross-Innes, C.S.; Becq, J.; Warren, A.; Cheetham, R.K.; Northen, H.; O’Donovan, M.; Malhotra, S.; di Pietro, M.; Ivakhno, S.; He, M.; et al. Whole-genome sequencing provides new insights into the clonal architecture of Barrett’s esophagus and esophageal adenocarcinoma. Nat. Genet. 2015, 47, 1038–1046. [Google Scholar] [CrossRef]
- Huang, H. Matrix Metalloproteinase-9 (MMP-9) as a Cancer Biomarker and MMP-9 Biosensors: Recent Advances. Sensors 2018, 18, 3249. [Google Scholar] [CrossRef] [Green Version]
- Lu, X.; Duan, L.; Xie, H.; Lu, X.; Lu, D.; Lu, D.; Jiang, N.; Chen, Y. Evaluation of MMP-9 and MMP-2 and their suppressor TIMP-1 and TIMP-2 in adenocarcinoma of esophagogastric junction. Oncol. Targets Ther. 2016, 9, 4343–4349. [Google Scholar]
- Herszenyi, L.; Hritz, I.; Pregun, I.; Sipos, F.; Juhasz, M.; Molnar, B.; Tulassay, Z. Alterations of glutathione S-transferase and matrix metalloproteinase-9 expressions are early events in esophageal carcinogenesis. World J. Gastroenterol. 2007, 13, 676–682. [Google Scholar] [CrossRef] [Green Version]
- Weh, K.M.; Turgeon, D.K.; Rubenstein, J.H.; Clarke, J.L.; Howell, A.B.; Chang, A.C.; Kresty, L.A. Proanthocyanidins mitigate bile acid-induced changes in GSTT2 levels in a panel of racially diverse patient-derived primary esophageal cell cultures. Mol. Carcinog. 2021, 61, 281–287. [Google Scholar] [CrossRef]
- Guo, J.C.; Xie, Y.M.; Ran, L.Q.; Cao, H.H.; Sun, C.; Wu, J.Y.; Wu, Z.Y.; Liao, L.D.; Zhao, W.J.; Fang, W.K.; et al. L1CAM drives oncogenicity in esophageal squamous cell carcinoma by stimulation of ezrin transcription. J. Mol. Med. 2017, 95, 1355–1368. [Google Scholar] [CrossRef]
- Chen, D.L.; Zeng, Z.L.; Yang, J.; Ren, C.; Wang, D.S.; Wu, W.J.; Xu, R.H. L1cam promotes tumor progression and metastasis and is an independent unfavorable prognostic factor in gastric cancer. J. Hematol. Oncol. 2013, 6, 43. [Google Scholar] [CrossRef] [Green Version]
- Ganesh, K.; Basnet, H.; Kaygusuz, Y.; Laughney, A.M.; He, L.; Sharma, R.; O’Rourke, K.P.; Reuter, V.P.; Huang, Y.H.; Turkekul, M.; et al. L1CAM defines the regenerative origin of metastasis-initiating cells in colorectal cancer. Nat. Cancer 2020, 1, 28–45. [Google Scholar] [CrossRef]
- Jacobi, N.; Seeboeck, R.; Hofmann, E.; Eger, A. ErbB Family Signalling: A Paradigm for Oncogene Addiction and Personalized Oncology. Cancers 2017, 9, 33. [Google Scholar] [CrossRef] [Green Version]
- Kumagai, S.; Koyama, S.; Nishikawa, H. Antitumour immunity regulated by aberrant ERBB family signalling. Nat. Rev. Cancer 2021, 21, 181–197. [Google Scholar] [CrossRef]
- Chen, J.; Zhou, J.; Gao, Z.; Li, X.; Wang, F.; Duan, X.; Li, G.; Joshi, B.P.; Kuick, R.; Appelman, H.D.; et al. Multiplexed Targeting of Barrett’s Neoplasia with a Heterobivalent Ligand: Imaging Study on Mouse Xenograft in Vivo and Human Specimens ex Vivo. J. Med. Chem. 2018, 61, 5323–5331. [Google Scholar] [CrossRef]
- Joshi, B.P.; Zhou, J.; Pant, A.; Duan, X.; Zhou, Q.; Kuick, R.; Owens, S.R.; Appelman, H.; Wang, T.D. Design and Synthesis of Near-Infrared Peptide for in Vivo Molecular Imaging of HER2. Bioconjug. Chem. 2016, 27, 481–494. [Google Scholar] [CrossRef] [Green Version]
- Kashyap, M.K.; Abdel-Rahman, O. Expression, regulation and targeting of receptor tyrosine kinases in esophageal squamous cell carcinoma. Mol. Cancer 2018, 17, 54. [Google Scholar] [CrossRef]
- Caspa Gokulan, R.; Garcia-Buitrago, M.T.; Zaika, A.I. From genetics to signaling pathways: Molecular pathogenesis of esophageal adenocarcinoma. Biochim. Biophys. Acta Rev. Cancer 2019, 1872, 37–48. [Google Scholar]
- Chiba, T. What are the real roles of different erbB proteins in Barrett’s Esophagus. Digestion 2004, 70, 93–94. [Google Scholar] [CrossRef]
- Peng, D.; Zaika, A.; Que, J.; El-Rifai, W. The antioxidant response in Barrett’s tumorigenesis: A double-edged sword. Redox. Biol. 2021, 41, 101894. [Google Scholar] [CrossRef]
- Suraweera, T.L.; Rupasinghe, H.P.V.; Dellaire, G.; Xu, Z. Regulation of Nrf2/ARE Pathway by Dietary Flavonoids: A Friend or Foe for Cancer Management? Antioxidants 2020, 9, 973. [Google Scholar] [CrossRef]
- Tao, S.; Rojo de la Vega, M.; Chapman, E.; Ooi, A.; Zhang, D.D. The effects of NRF2 modulation on the initiation and progression of chemically and genetically induced lung cancer. Mol. Carcinog. 2018, 57, 182–192. [Google Scholar] [CrossRef]
- Peng, D.; Lu, H.; Hu, T.; Sriramajayam, K.; El-Rifai, W. Abstract 1938: Targeting constitutively overexpressed NRF2 in esophageal adenocarcinoma. Cancer Res. 2020, 80, 1938. [Google Scholar]
- LaPlante, K.L.; Sarkisian, S.A.; Woodmansee, S.; Rowley, D.C.; Seeram, N.P. Effects of cranberry extracts on growth and biofilm production of Escherichia coli and Staphylococcus species. Phytother. Res. 2012, 26, 1371–1374. [Google Scholar] [CrossRef]
- Huang, Y.; Nikolic, D.; Pendland, S.; Doyle, B.J.; Locklear, T.D.; Mahady, G.B. Effects of cranberry extracts and ursolic acid derivatives on P-fimbriated Escherichia coli, COX-2 activity, pro-inflammatory cytokine release and the NF-kappabeta transcriptional response in vitro. Pharm. Biol. 2009, 47, 18–25. [Google Scholar] [CrossRef] [Green Version]
- Rane, H.S.; Bernardo, S.M.; Howell, A.B.; Lee, S.A. Cranberry-derived proanthocyanidins prevent formation of Candida albicans biofilms in artificial urine through biofilm- and adherence-specific mechanisms. J. Antimicrob. Chemother. 2014, 69, 428–436. [Google Scholar] [CrossRef] [Green Version]
- Philip, N.; Leishman, S.J.; Bandara, H.; Walsh, L.J. Polyphenol-Rich Cranberry Extracts Modulate Virulence of Streptococcus mutans-Candida albicans Biofilms Implicated in the Pathogenesis of Early Childhood Caries. Pediatr. Dent. 2019, 41, 56–62. [Google Scholar]
- Greene, A.C.; Acharya, A.P.; Lee, S.B.; Gottardi, R.; Zaleski, E.; Little, S.R. Cranberry extract-based formulations for preventing bacterial biofilms. Drug Deliv. Transl. Res. 2021, 11, 1144–1155. [Google Scholar] [CrossRef]










Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Weh, K.M.; Zhang, Y.; Howard, C.L.; Howell, A.B.; Clarke, J.L.; Kresty, L.A. Cranberry Polyphenols in Esophageal Cancer Inhibition: New Insights. Nutrients 2022, 14, 969. https://doi.org/10.3390/nu14050969
Weh KM, Zhang Y, Howard CL, Howell AB, Clarke JL, Kresty LA. Cranberry Polyphenols in Esophageal Cancer Inhibition: New Insights. Nutrients. 2022; 14(5):969. https://doi.org/10.3390/nu14050969
Chicago/Turabian StyleWeh, Katherine M., Yun Zhang, Connor L. Howard, Amy B. Howell, Jennifer L. Clarke, and Laura A. Kresty. 2022. "Cranberry Polyphenols in Esophageal Cancer Inhibition: New Insights" Nutrients 14, no. 5: 969. https://doi.org/10.3390/nu14050969
APA StyleWeh, K. M., Zhang, Y., Howard, C. L., Howell, A. B., Clarke, J. L., & Kresty, L. A. (2022). Cranberry Polyphenols in Esophageal Cancer Inhibition: New Insights. Nutrients, 14(5), 969. https://doi.org/10.3390/nu14050969