Associations between Frequency of Culinary Herb Use and Gut Microbiota
Abstract
:1. Introduction
1.1. Diet and the Microbiome
1.2. Culinary Herbs and Spices
1.3. Research Objective
2. Materials and Methods
2.1. Study Design
2.2. Participants
2.3. 16S rRNA Gene Sequencing and Processing
2.4. Microbial Ecology Analyses
2.5. Variables
2.5.1. Outcome Variables
2.5.2. Exposure Variables
2.5.3. Adjustment Variables
2.6. Statistical Models
3. Results
3.1. Descriptive Statistics
3.2. Frequency of Culinary Herb Use
3.3. Association between Frequency of Culinary Herb Use and Alpha Diversity
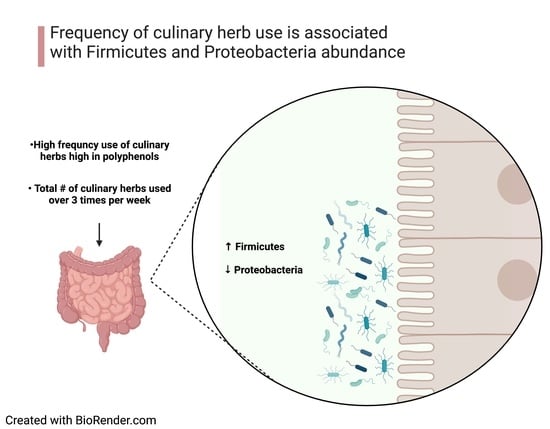
3.4. Association between Frequency of Culinary Herb Use and Phylum Abundance
4. Discussion
Strengths and Limitations
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Turnbaugh, P.J.; Ley, R.E.; Hamady, M.; Fraser-Liggett, C.M.; Knight, R.; Gordon, J.I. The Human Microbiome Project. Nature 2007, 449, 804–810. [Google Scholar] [CrossRef] [PubMed]
- Ursell, L.K.; Metcalf, J.L.; Parfrey, L.W.; Knight, R. Defining the human microbiome. Nutr. Rev. 2012, 70, S38–S44. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Amon, P.; Sanderson, I. What is the microbiome? Arch. Dis. Child. Educ. Pract. Ed. 2017, 102, 258–261. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, G.D.; Chen, J.; Hoffmann, C.; Bittinger, K.; Chen, Y.Y.; Keilbaugh, S.A.; Bewtra, M.; Knights, D.; Walters, W.A.; Knight, R.; et al. Linking long-term dietary patterns with gut microbial enterotypes. Science 2011, 334, 105–108. [Google Scholar] [CrossRef] [Green Version]
- Flint, H.J.; Duncan, S.H.; Louis, P. The impact of nutrition on intestinal bacterial communities. Curr. Opin. Microbiol. 2017, 38, 59–65. [Google Scholar] [CrossRef]
- Heintz-Buschart, A.; Wilmes, P. Human Gut Microbiome: Function Matters. Trends Microbiol. 2018, 26, 563–574. [Google Scholar] [CrossRef]
- Clemente, J.C.; Manasson, J.; Scher, J.U. The role of the gut microbiome in systemic inflammatory disease. BMJ 2018, 360, j5145. [Google Scholar] [CrossRef]
- Holscher, H.D. Dietary fiber and prebiotics and the gastrointestinal microbiota. Gut Microbes 2017, 8, 172–184. [Google Scholar] [CrossRef]
- Biesalski, H.K. Nutrition meets the microbiome: Micronutrients and the microbiota. Ann. N. Y. Acad. Sci. 2016, 1372, 53–64. [Google Scholar] [CrossRef]
- Vallverdú-Queralt, A.; Regueiro, J.; Martínez-Huélamo, M.; Rinaldi Alvarenga, J.F.; Leal, L.N.; Lamuela-Raventos, R.M. A comprehensive study on the phenolic profile of widely used culinary herbs and spices: Rosemary, thyme, oregano, cinnamon, cumin and bay. Food Chem. 2014, 154, 299–307. [Google Scholar] [CrossRef]
- Chandrababu, S.; Bastola, D. An Integrated Approach to Recognize Potential Protective Effects of Culinary Herbs against Chronic Diseases. J. Healthc. Inform. Res. 2019, 3, 184–199. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Liang, Q.; Balakrishnan, B.; Belobrajdic, D.P.; Feng, Q.J.; Zhang, W. Role of dietary nutrients in the modulation of gut microbiota: A narrative review. Nutrients 2020, 12, 381. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alves-Santos, A.M.; Sugizaki, C.S.A.; Lima, G.C.; Naves, M.M.V. Prebiotic effect of dietary polyphenols: A systematic review. J. Funct. Foods 2020, 74, 104169. [Google Scholar] [CrossRef]
- Opara, E.I.; Chohan, M. Culinary herbs and spices: Their bioactive properties, the contribution of polyphenols and the challenges in deducing their true health benefits. Int. J. Mol. Sci. 2014, 15, 19183–19202. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mukherjee, C.; Chakraborty, S. Study of dietary polyphenols from natural herbal sources for providing protection against human degenerative disorders. Biocatal. Agric. Biotechnol. 2021, 33, 101956. [Google Scholar] [CrossRef]
- Ma, Z.J.; Wang, H.J.; Ma, X.J.; Li, Y.; Yang, H.J.; Li, H.; Su, J.R.; Zhang, C.E.; Huang, L.Q. Modulation of gut microbiota and intestinal barrier function during alleviation of antibiotic-associated diarrhea with Rhizoma: Zingiber officinale (Ginger) extract. Food Funct. 2020, 11, 10839–10851. [Google Scholar] [CrossRef]
- Rosca, A.E.; Iesanu, M.I.; Zahiu, C.D.M.; Voiculescu, S.E.; Paslaru, A.C.; Zagrean, A.M. Capsaicin and gut microbiota in health and disease. Molecules 2020, 25, 5681. [Google Scholar] [CrossRef]
- Thompson, A.; Meah, D.; Ahmed, N.; Conniff-Jenkins, R.; Chileshe, E.; Phillips, C.O.; Claypole, T.C.; Forman, D.W.; Row, P.E. Comparison of the antibacterial activity of essential oils and extracts of medicinal and culinary herbs to investigate potential new treatments for irritable bowel syndrome. BMC Complement. Altern. Med. 2013, 13, 338. [Google Scholar] [CrossRef] [Green Version]
- Lu, Q.Y.; Summanen, P.H.; Lee, R.P.; Huang, J.; Henning, S.M.; Heber, D.; Finegold, S.M.; Li, Z. Prebiotic Potential and Chemical Composition of Seven Culinary Spice Extracts. J. Food Sci. 2017, 82, 1807–1813. [Google Scholar] [CrossRef] [Green Version]
- Khine, W.W.T.; Haldar, S.; De Loi, S.; Lee, Y.K. A single serving of mixed spices alters gut microflora composition: A dose–response randomised trial. Sci. Rep. 2021, 11, 1–12. [Google Scholar] [CrossRef]
- Lu, Q.Y.; Rasmussen, A.M.; Yang, J.; Lee, R.P.; Huang, J.; Shao, P.; Carpenter, C.; Gilbuena, I.; Thames, G.; Henning, S.; et al. Mixed spices at culinary doses have prebiotic effects in healthy adults: A pilot study. Nutrients 2019, 11, 1425. [Google Scholar] [CrossRef] [Green Version]
- Ried, K.; Travica, N.; Dorairaj, R.; Sali, A. Herbal formula improves upper and lower gastrointestinal symptoms and gut health in Australian adults with digestive disorders. Nutr. Res. 2020, 76, 37–51. [Google Scholar] [CrossRef] [PubMed]
- Ghitea, T.C.; El-Kharoubi, A.; Ganea, M.; Bimbo-Szuhai, E.; Nemeth, T.S.; Ciavoi, G.; Foghis, M.; Dobjanschi, L.; Pallag, A.; Micle, O. The antimicrobial activity of origanum vulgare l. Correlated with the gastrointestinal perturbation in patients with metabolic syndrome. Molecules 2021, 26, 283. [Google Scholar] [CrossRef] [PubMed]
- Kang, C.; Zhang, Y.; Zhu, X.; Liu, K.; Wang, X.; Chen, M.; Wang, J.; Chen, H.; Hui, S.; Huang, L.; et al. Healthy subjects differentially respond to dietary capsaicin correlating with specific gut enterotypes. J. Clin. Endocrinol. Metab. 2016, 101, 4681–4689. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lopresti, A.L.; Smith, S.J.; Rea, A.; Michel, S. Efficacy of a curcumin extract (CurcugenTM) on gastrointestinal symptoms and intestinal microbiota in adults with self-reported digestive complaints: A randomised, double-blind, placebo-controlled study. BMC Complement. Med. Ther. 2021, 21, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Ghaisas, S.; Maher, J.; Kanthasamy, A. Gut microbiome in health and disease: Linking the microbiome-gut-brain axis and environmental factors in the pathogenesis of systemic and neurodegenerative diseases. Pharmacol. Ther. 2016, 158, 52–62. [Google Scholar] [CrossRef] [Green Version]
- Bradley, R.; Pickworth, C.K.; Wexler, R.S.; Sadowski, A.; Buttolph, L.; Sarrar, H.; Moehle, J.; Torrens, M.T.; Harnett, J.; McIntyre, E.; et al. Protocol for the international cohort on lifestyle determinants of health study: A longitudinal investigation of complementary and integrative health utilization in postsecondary education students. J. Altern. Complement. Med. 2021, 27, 184–191. [Google Scholar] [CrossRef]
- Harris, P.A.; Taylor, R.; Thielke, R.; Payne, J.; Gonzalez, N.; Conde, J.G. Research electronic data capture (REDCap)-A metadata-driven methodology and workflow process for providing translational research informatics support. J. Biomed. Inform. 2009, 42, 377–381. [Google Scholar] [CrossRef] [Green Version]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [Green Version]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef] [PubMed]
- R Development Core Team. R: A Language and Environment for Statistical Computing; Version 4.0.5; R Foundation for Statistical Computing: Vienna, Austria, 2021; ISBN 3-900051-07-0. [Google Scholar]
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis; Springer: Berlin/Heidelberg, Germany, 2016. [Google Scholar]
- Shannon, C.E. A mathematical theory of communication. Bell Syst. Tech. J. 1948, 27, 379–423. [Google Scholar] [CrossRef] [Green Version]
- Oksanen, J.; Blanchet, F.G.; Friendly, M.; Kindt, R.; Legendre, P.; Mcglinn, D.; Minchin, P.R.; O’hara, R.B.; Simpson, G.L.; Solymos, P.; et al. Vegan: Community Ecology Package. R Package Version 2.5-5. 2019. Available online: https://CRAN.R-project.org/package=vegan (accessed on 17 April 2022).
- Paradis, E.; Schliep, K. Ape 5.0: An environment for modern phylogenetics and evolutionary analyses in R. Bioinformatics 2019, 35, 526–528. [Google Scholar] [CrossRef] [PubMed]
- U.S. Department of Agriculture, Agricultural Research Service. Dr. Duke’s Phytochemical and Ethnobotanical Databases; USDA: Washington, DC, USA, 1992.
- Neveu, V.; Perez-Jiménez, J.; Vos, F.; Crespy, V.; du Chaffaut, L.; Mennen, L.; Knox, C.; Eisner, R.; Cruz, J.; Wishart, D.; et al. Phenol-Explorer: An online comprehensive database on polyphenol contents in foods. Database 2010, 2010, bap024. [Google Scholar] [CrossRef]
- Rothwell, J.A.; Perez-Jimenez, J.; Neveu, V.; Medina-Remón, A.; M’Hiri, N.; García-Lobato, P.; Manach, C.; Knox, C.; Eisner, R.; Wishart, D.S.; et al. Phenol-Explorer 3.0: A major update of the Phenol-Explorer database to incorporate data on the effects of food processing on polyphenol content. Database 2013, 2013, bat070. [Google Scholar] [CrossRef]
- IBM Corporation. IBM SPSS Statistics 28.0 for Macintosh; IBM Corporation: Armonk, NY, USA, 2021; Volume 2021. [Google Scholar]
- Mostafa, I.; Abbas, H.A.; Ashour, M.L.; Yasri, A.; El-Shazly, A.M.; Wink, M.; Sobeh, M. Polyphenols from salix tetrasperma impair virulence and inhibit quorumsensing of pseudomonas aeruginosa. Molecules 2020, 25, 1341. [Google Scholar] [CrossRef] [Green Version]
- Nazareth, M.S.; Shreelakshmi, S.V.; Shetty, N.P. Identification and Characterization of Polyphenols from Carissa spinarum Fruit and Evaluation of Their Antioxidant and Anti-quorum Sensing Activity. Curr. Microbiol. 2021, 78, 1277–1285. [Google Scholar] [CrossRef]
- Liu, M.; Amini, A.; Ahmad, Z. Safranal and its analogs inhibit Escherichia coli ATP synthase and cell growth. Int. J. Biol. Macromol. 2017, 95, 145–152. [Google Scholar] [CrossRef] [Green Version]
- Dadi, P.K.; Ahmad, M.; Ahmad, Z. Inhibition of ATPase activity of Escherichia coli ATP synthase by polyphenols. Int. J. Biol. Macromol. 2009, 45, 72–79. [Google Scholar] [CrossRef]
- Nakanishi-Matsui, M.; Sekiya, M.; Futai, M. ATP synthase from Escherichia coli: Mechanism of rotational catalysis, and inhibition with the ε subunit and phytopolyphenols. Biochim. Biophys. Acta Bioenergy 2016, 1857, 129–140. [Google Scholar] [CrossRef]
- Peron, G.; Gargari, G.; Meroño, T.; Miñarro, A.; Lozano, E.V.; Escuder, P.C.; González-Domínguez, R.; Hidalgo-Liberona, N.; Del Bo, C.; Bernardi, S.; et al. Crosstalk among intestinal barrier, gut microbiota and serum metabolome after a polyphenol-rich diet in older subjects with “leaky gut”: The MaPLE trial. Clin. Nutr. 2021, 40, 5288–5297. [Google Scholar] [CrossRef] [PubMed]
- Vetrani, C.; Maukonen, J.; Bozzetto, L.; Della Pepa, G.; Vitale, M.; Costabile, G.; Riccardi, G.; Rivellese, A.A.; Saarela, M.; Annuzzi, G. Diets naturally rich in polyphenols and/or long-chain n-3 polyunsaturated fatty acids differently affect microbiota composition in high-cardiometabolic-risk individuals. Acta Diabetol. 2020, 57, 853–860. [Google Scholar] [CrossRef] [PubMed]
- Moreno-Indias, I.; Sánchez-Alcoholado, L.; Pérez-Martínez, P.; Andrés-Lacueva, C.; Cardona, F.; Tinahones, F.; Queipo-Ortuño, M.I. Red wine polyphenols modulate fecal microbiota and reduce markers of the metabolic syndrome in obese patients. Food Funct. 2016, 7, 1775–1787. [Google Scholar] [CrossRef] [Green Version]
- Bauer, B.W.; Gangadoo, S.; Bajagai, Y.S.; Van, T.T.H.; Moore, R.J.; Stanley, D. Oregano powder reduces Streptococcus and increases SCFA concentration in a mixed bacterial culture assay. PLoS ONE 2019, 14, e0216853. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ruan, D.; Fan, Q.; Fouad, A.M.; Sun, Y.; Huang, S.; Wu, A.; Lin, C.; Kuang, Z.; Zhang, C.; Jiang, S. Effects of dietary oregano essential oil supplementation on growth performance, intestinal antioxidative capacity, immunity, and intestinal microbiota in yellow-feathered chickens. J. Anim. Sci. 2021, 99, skab033. [Google Scholar] [CrossRef] [PubMed]
- Hussein, M.M.A.; Abd El-Hack, M.E.; Mahgoub, S.A.; Saadeldin, I.M.; Swelum, A.A. Effects of clove (Syzygium aromaticum) oil on quail growth, carcass traits, blood components, meat quality, and intestinal microbiota. Poult. Sci. 2019, 98, 319–329. [Google Scholar] [CrossRef]
- Qi, L.; Mao, H.; Lu, X.; Shi, T.; Wang, J. Cinnamaldehyde Promotes the Intestinal Barrier Functions and Reshapes Gut Microbiome in Early Weaned Rats. Front. Nutr. 2021, 8, 772. [Google Scholar] [CrossRef]
- Mooyottu, S.; Flock, G.; Upadhyay, A.; Upadhyaya, I.; Maas, K.; Venkitanarayanan, K. Protective effect of carvacrol against gut dysbiosis and Clostridium difficile associated disease in a mouse model. Front. Microbiol. 2017, 8, 625. [Google Scholar] [CrossRef]
- Silva Júnior, C.D.; Martins, C.C.S.; Dias, F.T.F.; Sitanaka, N.Y.; Ferracioli, L.B.; Moraes, J.E.; Pizzolante, C.C.; Budiño, F.E.L.; Pereira, R.; Tizioto, P.; et al. The use of an alternative feed additive, containing benzoic acid, thymol, eugenol, and piperine, improved growth performance, nutrient and energy digestibility, and gut health in weaned piglets. J. Anim. Sci. 2020, 98, skaa119. [Google Scholar] [CrossRef]
- McDonald, D.; Hyde, E.; Debelius, J.W.; Morton, J.T.; Gonzalez, A.; Ackermann, G.; Aksenov, A.A.; Behsaz, B.; Brennan, C.; Chen, Y.; et al. American Gut: An Open Platform for Citizen Science Microbiome Research. mSystems 2018, 3, e00031-18. [Google Scholar] [CrossRef] [Green Version]
- Kashtanova, D.; Tkacheva, O.; Popenko, A.; Egshatyan, L.; Tyakht, A.; Alexeev, D.; Kotovskaya, Y.; Plokhova, E.; Boytsov, S. Gut microbiota and vascular biomarkers in patients without clinical cardiovascular diseases. Artery Res. 2017, 18, 41–48. [Google Scholar] [CrossRef]
- Sims, T.T.; Colbert, L.E.; Zheng, J.; Delgado Medrano, A.Y.; Hoffman, K.L.; Ramondetta, L.; Jazaeri, A.; Jhingran, A.; Schmeler, K.M.; Daniel, C.R.; et al. Gut microbial diversity and genus-level differences identified in cervical cancer patients versus healthy controls. Gynecol. Oncol. 2019, 155, 237–244. [Google Scholar] [CrossRef] [PubMed]
- Turnbaugh, P.J.; Hamady, M.; Yatsunenko, T.; Cantarel, B.L.; Duncan, A.; Ley, R.E.; Sogin, M.L.; Jones, W.J.; Roe, B.A.; Affourtit, J.P.; et al. A core gut microbiome in obese and lean twins. Nature 2009, 457, 480–484. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Menni, C.; Jackson, M.A.; Pallister, T.; Steves, C.J.; Spector, T.D.; Valdes, A.M. Gut microbiome diversity and high-fibre intake are related to lower long-term weight gain. Int. J. Obes. 2017, 41, 1099–1105. [Google Scholar] [CrossRef] [PubMed] [Green Version]


| Variables | Value |
|---|---|
| Alpha Diversity | M (SD) |
| Shannon Index | 29.34 (6.13) |
| Phylum Abundance | M (SD) |
| Firmicutes | 2837.28 (398.21) |
| Bacteroidota | 738.44 (309.27) |
| Actinobacteria | 150.59 (172.71) |
| Proteobacteria | 181.93 (243.70) |
| Frequency of Herb Use | M (SD) |
| Allium | 3.86 (2.16) |
| Eugenol | 1.87 (1.67) |
| Capsaicin | 2.68 (1.42) |
| >30,000 PPM Polyphenol | 2.43 (1.40) |
| >50,000 PPM Polyphenol | 2.40 (1.54) |
| >30,000 PPM Antibiotic | 2.07 (0.33) |
| >90,000 PPM Antibiotic | 2.55 (1.44) |
| Variable | Value |
|---|---|
| Age | M (SD) |
| 29.34 (6.13) | |
| Sex Assigned at Birth | n (%) |
| Male | 14 (14.6%) |
| Female | 81 (84.4%) |
| Intersex | 1 (>1%) |
| Race | n(%) |
| White/Caucasian | 75 (78.1%) |
| Asian | 5 (5.2%) |
| African American | 2 (2%) |
| Middle Eastern | 2 (2%) |
| Native Hawaiian/Pacific Islander | 1 (1%) |
| American Native/Alaska Native | 1 (1%) |
| Mixed | 6 (6.3%) |
| Other/Unknown | 4 (4.2%) |
| Ethnicity | n (%) |
| Hispanic/LatinX | 9 (9.4%) |
| Non-Hispanic/LatinX | 83 (86.5%) |
| Unknown | 4 (4.2%) |
| Dietary Factors | M (SD) |
| Est. daily fat intake (g) | 80.6 (37.5) |
| Est. daily protein intake (g) | 64.7 (30.7) |
| Est. daily fiber intake (g) | 27.2 (10.8) |
| Medications | M (SD) |
| Total Used | 0.6 (1.0) |
| Supplements | M (SD) |
| Total Used | 5.7 (4.9) |
| Exposure: | Allium | Capsaicin | Eugenol | Antibiotic | Antibiotic | Polyphenol | Polyphenol |
|---|---|---|---|---|---|---|---|
| Freq. of Use | >30,000 PPM | >90,000 PPM | >30,000 PPM | >50,000 PPM | |||
| β (CI) | β (CI) | β (CI) | β (CI) | β (CI) | β (CI) | β (CI) | |
| p-Value | p-Value | p-Value | p-Value | p-Value | p-Value | p-Value | |
| Model 1 | −0.067 | 0.009 | 0.015 | 0.079 | 0.136 | 0.056 | −0.054 |
| (−0.068, 0.035) | (−0.063, 0.068) | (−0.054, 0.062) | (−0.037, 0.082) | (−0.021, 0.107) | (−0.047, 0.83) | (−0.076, 0.044) | |
| 0.517 | 0.931 | 0.888 | 0.446 | 0.187 | 0.587 | 0.6 | |
| Model 2 | −0.042 | 0.018 | 0.039 | 0.104 | 0.135 | 0.048 | −0.042 |
| (−0.065, 0.043) | (−0.059, 0.071) | (−0.048, 0.070) | (−0.030, 0.090) | (−0.021, 0.106) | (−0.050, 0.082) | (−0.073, 0.049) | |
| 0.696 | 0.858 | 0.709 | 0.32 | 0.189 | 0.646 | 0.69 | |
| Model 3 | −0.055 | −0.078 | −0.048 | 0.063 | 0.109 | 0.044 | −0.109 |
| (−0.068, 0.041) | (−0.094, 0.045) | (−0.076, 0.049) | (−0.047, 0.0.79) | (−0.031, 0.099) | (−0.057, 0.085) | (−0.098, 0.033) | |
| 0.614 | 0.477 | 0.667 | 0.576 | 0.296 | 0.693 | 0.33 | |
| Model 4 | −0.045 | −0.094 | −0.041 | 0.066 | 0.119 | 0.036 | −0.109 |
| (−0.067, 0.044) | (−0.101, 0.041) | (−0.077, 0.054) | (−0.047, 0.063) | (−0.029, 0.104) | (−0.062, 0.085) | (−0.099, 0.035) | |
| 0.688 | 0.403 | 0.725 | 0.564 | 0.266 | 0.754 | 0.342 |
| Exposure: | Shannon Index | Firmicutes | Bacteroidota | Proteobacteria | Actinobacteria |
|---|---|---|---|---|---|
| Freq. of Use | |||||
| β (CI) | β (CI) | β (CI) | β (CI) | β (CI) | |
| p-Value | p-Value | p-Value | p-Value | p-Value | |
| Model 1 | 0.042 | 0.289 | −0.154 | −0.194 | −0.037 |
| (−0.012, 0.018) | (11.2, 59.2) | (−33.8, 4.6) | (−29.5, 0.572) | (−12.8, 8.9) | |
| 0.684 | 0.004 ** | 0.135 | 0.059 | 0.725 | |
| Model 2 | 0.046 | 0.286 | −0.153 | −0.198 | −0.029 |
| (−0.012, 0.018) | (9.50, 59.7) | (−34.0, 5.8) | (−30.4, 0.940) | (−12.6, 9.0) | |
| 0.66 | 0.007 ** | 0.152 | 0.064 | 0.783 | |
| Model 3 | −0.022 | 0.301 | −0.137 | −0.235 | −0.015 |
| (−0.018, 0.014) | (9.0, 63.9) | (−34.1, 9.8) | (−34.9, −0.590) | (−13.3, 10.5) | |
| 0.842 | 0.009 ** | 0.243 | 0.044 * | 0.897 | |
| Model 4 | −0.021 | 0.294 | −0.145 | −0.216 | −0.005 |
| (−0.018, 0.015) | (8.4, 62.2) | (−35.3, 9.5) | (−32.8, 0.462) | (−13.0, 11.2) | |
| 0.855 | 0.009 ** | 0.223 | 0.055 | 0.968 |
| Exposure: | Allium | Capsaicin | Eugenol | Antibiotic | Antibiotic | Polyphenol | Polyphenol |
|---|---|---|---|---|---|---|---|
| Freq. of Use | >30,00 PPM | >30,000 PPM | >90,000 PPM | >50,000 PPM | |||
| β (CI) | β (CI) | β (CI) | β (CI) | β (CI) | β (CI) | β (CI) | |
| p-Value | p-Value | p-Value | p-Value | p-Value | p-Value | p-Value | |
| Model 1 | −0.044 | −0.017 | −0.033 | −0.077 | −0.102 | −0.119 | 0.032 |
| (−46.61, 30.06) | (−52.64, 44.46) | (−48.98, 36.55) | (−60.72, 27.69) | (−71.35, 24.04) | (−77.05, 20.34) | (−37.79, 51.77) | |
| 0.669 | 0.867 | 0.773 | 0.46 | 0.327 | 0.251 | 0.757 | |
| Model 2 | 0.01 | −0.015 | 0.039 | −0.03 | −0.107 | −0.09 | 0.059 |
| (−37.12, 40.80) | (−50.68, 43.68) | (−35.09, 51.13) | (−50.23, 37.31) | (−71.48, 21.53) | (−69.94, 26.03) | (−31.25, 57.07) | |
| 0.926 | 0.883 | 0.713 | 0.771 | 0.289 | 0.379 | 0.563 | |
| Model 3 | −0.001 | 0.01 | 0.071 | −0.031 | −0.112 | −0.12 | 0.072 |
| (−40.83, 40.40) | (−49.57, 54.39) | (−32.31, 61.68) | (−55.23, 41.75) | (−74.56, 22.32) | (−83.90, 21.70) | (−33.30, 64.43) | |
| 0.992 | 0.927 | 0.536 | 0.783 | 0.287 | 0.287 | 0.528 | |
| Model 4 | −0.002 | 0.024 | 0.095 | −0.033 | −0.105 | −0.106 | 0.083 |
| (−41.70, 40.99) | (−47.70, 58.99) | (−29.78, 69.31) | (−55.46, 42.46) | (−74.38, 25.54) | (−82.87, 26.36) | (−31.77, 67.81) | |
| 0.986 | 0.834 | 0.43 | 0.792 | 0.334 | 0.36 | 0.474 |
| Exposure: | Allium | Capsaicin | Eugenol | Antibiotic | Antibiotic | Polyphenol | Polyphenol |
|---|---|---|---|---|---|---|---|
| Freq. of Use | >30,000 PPM | >90,000 PPM | >30,000 PPM | >50,000 PPM | |||
| β (CI) | β (CI) | β (CI) | β (CI) | β (CI) | β (CI) | β (CI) | |
| p-Value | p-Value | p-Value | p-Value | p-Value | p-Value | p-Value | |
| Model 1 | −0.067 | −0.164 | −0.046 | −0.135 | −0.087 | −0.13 | −0.067 |
| (−81.07, 56.26) | (−155.98, 16.58) | (−93.79, 59.26) | (−126.90, 30.64) | (−121.66, 49.39) | (−142.59, 31.56) | (−106.24, 53.87) | |
| 0.517 | 0.113 | 0.655 | 0.191 | 0.404 | 0.209 | 0.518 | |
| Model 2 | −0.042 | −0.171 | −0.054 | −0.124 | −0.074 | −0.125 | −0.061 |
| (−92.79, 51.68) | (−158.38, 14.22) | (−100.00, 60.02) | (−132.96, 28.18) | (−117.66, 55.68) | (−141.26, 36.43) | (−105.67, 58.35) | |
| 0.696 | 0.101 | 0.621 | 0.253 | 0.497 | 0.238 | 0.568 | |
| Model 3 | −0.055 | −0.157 | −0.015 | −0.106 | −0.053 | −0.118 | −0.025 |
| (−89.54, 61.65) | (−161.90, 29.66) | (−93.28, 82.15) | (−136.83, 42.75) | (−112.84, 68.56) | (−145.72, 51.50) | (−101.10, 81.31) | |
| 0.614 | 0.174 | 0.9 | 0.364 | 0.629 | 0.31 | 0.83 | |
| Model 4 | −0.045 | −0.16 | −0.032 | −0.128 | −0.064 | −0.127 | −0.032 |
| (−93.48, 60.67) | (−165.91, 31.08) | (−104.71, 80.83) | (−138.61, 43.00) | (−120.34, 66.80) | (−152.21, 51.70) | (−105.68, 80.64) | |
| 0.688 | 0.177 | 0.799 | 0.298 | 0.571 | 0.292 | 0.79 |
| Exposure: | Allium. | Capsaicin | Eugenol | Antibiotic | Antibiotic | Polyphenol | Polyphenol |
|---|---|---|---|---|---|---|---|
| Freq. of Use | >30,000 PPM | >90,000 PPM | >30,000 PPM | >50,000 PPM | |||
| β (CI) | β (CI) | β (CI) | β (CI) | β (CI) | β (CI) | β (CI) | |
| p-Value | p-Value | p-Value | p-Value | p-Value | p-Value | p-Value | |
| Model 1 | 0.113 | 0.195 | 0.127 | 0.198 | 0.347 | 0.255 | 0.279 |
| (−39.3, 136.6) | (−3.5, 216.1) | (−37.2, 158.5) | (−1.7, 198.7) | (78.2, 275.0) | (26.3, 214.7) | (44.8, 261.9) | |
| 0.275 | 0.058 | 0.221 | 0.054 | 0.001 ** | 0.013 * | 0.006 ** | |
| Model 2 | 0.106 | 0.198 | 0.114 | 0.192 | 0.342 | 0.247 | 0.272 |
| (−48.2, 139.2) | (−4.3, 219.2) | (−48.8, 158.4) | (−8.5, 199.3) | (71.4, 276.6) | (19.1, 214.4) | (37.4, 261.8) | |
| 0.337 | 0.059 | 0.296 | 0.071 | 0.001 ** | 0.020 * | 0.010 * | |
| Model 3 | 0.101 | 0.221 | 0.084 | 0.193 | 0.33 | 0.284 | 0.327 |
| (−53.9, 140.6) | (−18.3, 227.7) | (−72.6, 153.3) | (−4.3, 224.2) | (61.8, 274.4) | (27.6, 241.2) | (57.7, 301.8) | |
| 0.378 | 0.059 | 0.479 | 0.094 | 0.002 ** | 0.014 * | 0.004 ** | |
| Model 4 | 0.131 | 0.142 | 0.089 | 0.225 | 0.302 | 0.266 | 0.286 |
| (−38.7, 151.0) | (−44.9, 199.6) | (−72.2, 157.1) | (0.86, 222.2) | (46.0, 262.1) | (18.4, 233.2) | (34.6, 279.5) | |
| 0.242 | 0.212 | 0.463 | 0.048 * | 0.006 ** | 0.022 * | 0.013 * |
| Exposure: | Allium | Capsaicin | Eugenol | Antibiotic | Antibiotic | Polyphenol | Polyphenol |
|---|---|---|---|---|---|---|---|
| Freq. of Use | >30,000 PPM | >90,000 PPM | >30,000 PPM | >50,000 PPM | |||
| β (CI) | β (CI) | β (CI) | β (CI) | β (CI) | β (CI) | β (CI) | |
| p-Value | p-Value | p-Value | p-Value | p-Value | p-Value | p-Value | |
| Model 1 | −0.057 | −0.091 | −0.102 | −0.141 | −0.347 | −0.193 | −0.258 |
| (−69.05, 39.07) | (−98.67, 37.79) | (−89.96, 30.15) | (−104.81, 19.05) | (−143.73, −19.82) | (−132.80, 2.99) | (−140.21, −18.03) | |
| 0.583 | 0.378 | 0.325 | 0.072 | 0.010 ** | 0.061 | 0.012 * | |
| Model 2 | −0.037 | −0.087 | −0.099 | −0.137 | −0.342 | −0.203 | −0.262 |
| (−67.29, 47.57) | (−98.39, 40.25) | (−92.27, 34.40) | (−105.61, 22.36) | (−147.92, −19.74) | (−137.90, 1.36) | (−143.44, −17.42) | |
| 0.734 | 0.407 | 0.366 | 0.199 | 0.011 * | 0.055 | 0.013 * | |
| Model 3 | −0.038 | −0.11 | −0.113 | −0.189 | −0.227 | −0.257 | −0.326 |
| (−70.01, 50.09) | (−113.10, 39.90) | (−120.49, 36.14) | (−128.23, 13.17) | (−152.76, −19.71) | (−164.08, −11.1) | (−169.30, −30.88) | |
| 0.742 | 0.344 | 0.344 | 0.109 | 0.012 * | 0.025 * | 0.005 ** | |
| Model 4 | −0.063 | −0.048 | −0.098 | −0.191 | −0.225 | −0.201 | −0.311 |
| (−74.68, 41.29) | (−90.86, 59.93) | (−98.41, 40.78) | (−125.60, 9.73) | (−137.05, −3.03) | (−143.45, 8.16) | (−162.44, −28.34) | |
| 0.568 | 0.673 | 0.413 | 0.092 | 0.041 * | 0.078 | 0.006 ** |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vita, A.A.; McClure, R.; Farris, Y.; Danczak, R.; Gundersen, A.; Zwickey, H.; Bradley, R. Associations between Frequency of Culinary Herb Use and Gut Microbiota. Nutrients 2022, 14, 1981. https://doi.org/10.3390/nu14091981
Vita AA, McClure R, Farris Y, Danczak R, Gundersen A, Zwickey H, Bradley R. Associations between Frequency of Culinary Herb Use and Gut Microbiota. Nutrients. 2022; 14(9):1981. https://doi.org/10.3390/nu14091981
Chicago/Turabian StyleVita, Alexandra Adorno, Ryan McClure, Yuliya Farris, Robert Danczak, Anders Gundersen, Heather Zwickey, and Ryan Bradley. 2022. "Associations between Frequency of Culinary Herb Use and Gut Microbiota" Nutrients 14, no. 9: 1981. https://doi.org/10.3390/nu14091981
APA StyleVita, A. A., McClure, R., Farris, Y., Danczak, R., Gundersen, A., Zwickey, H., & Bradley, R. (2022). Associations between Frequency of Culinary Herb Use and Gut Microbiota. Nutrients, 14(9), 1981. https://doi.org/10.3390/nu14091981