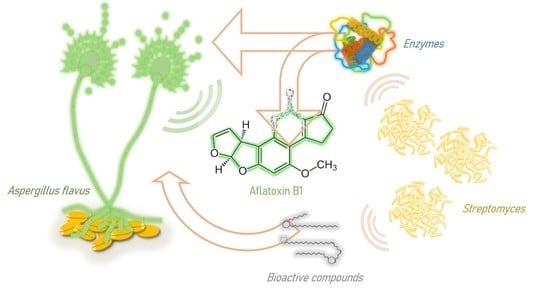
Aspergillus flavus Growth Inhibition and Aflatoxin B1 Decontamination by Streptomyces Isolates and Their Metabolites
Abstract
:1. Introduction
2. Results
2.1. Evaluation of the Antagonistic Activity of Streptomyces Isolates and Their Cell-Free Extracts (CFEs)
2.2. Mycotoxin Degradation Assay
2.3. Global Analysis of Strains: Cluster Analysis and Pearson Correlation Index
- -
- flavus growth in dual culture or with the addition of CFEs to the medium (Category 1);
- -
- AFB1-specific production by A. flavus during growth in dual culture or with CFEs added to the medium (Category 2);
- -
- AFB1 degradation by Streptomyces isolates in solid and liquid media, as well as by their CFEs (Category 3).
2.4. Search for Degradation By-Products
2.5. Evaluation of Residual Toxicity
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Streptomyces Isolates
5.2. Streptomyces Isolates Spore Numeration by Flux Cytometer
5.3. Pathogen Strains
5.4. Antagonistic Evaluation In Vitro
5.4.1. Streptomyces Isolate Cells in Dual Cultures on Solid Medium
5.4.2. Cell-Free Extracts of Liquid Cultures
5.4.3. Pathogen Surface Growth Measurement and AFB1-Specific Production Calculation
5.5. Mycotoxin Degradation Assay
5.5.1. Screening of AFB1 Degradation by Streptomyces Isolates and Their CFEs
5.5.2. Degradation Assays for the Search of Breakdown Products
5.6. Mycotoxin and Degradation By-Products Analysis
5.6.1. Sample Extraction
5.6.2. AFB1 Quantification by Fluorescence HPLC
5.6.3. Search for Degradation By-Products by HPLC-MS
5.7. Evaluation of Residual Toxicity by SOS Chromotest
- absorbance at 600 nm for sample wells;
- average absorbance at 600 nm for reagent blank wells;
- absorbance at 600 nm for negative control wells;
- absorbance at 420 nm for sample wells;
- average absorbance at 420 nm for reagent blank wells;
- absorbance at 420 nm for negative control wells.
5.8. Data Analysis and Data Visualization
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Oliveira, C.A.F.; Bovo, F.; Humberto, C.; Vincenzi, A.; Ravindranadha, K. Recent Trends in Microbiological Decontamination of Aflatoxins in Foodstuffs. In Aflatoxins—Recent Advances and Future Prospects; InTech: Rijeka, Croatia, 2013. [Google Scholar]
- McCormick, S.P. Microbial Detoxification of Mycotoxins. J. Chem. Ecol. 2013, 39, 907–918. [Google Scholar] [CrossRef] [PubMed]
- Riba, A.; Bouras, N.; Mokrane, S.; Mathieu, F.; Lebrihi, A.; Sabaou, N. Aspergillus section Flavi and aflatoxins in Algerian wheat and derived products. Food Chem. Toxicol. 2010, 48, 2772–2777. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Masood, M.; Iqbal, S.Z.; Asi, M.R.; Malik, N. Natural occurrence of aflatoxins in dry fruits and edible nuts. Food Control 2015, 55, 62–65. [Google Scholar] [CrossRef]
- Azzoune, N.; Mokrane, S.; Riba, A.; Bouras, N.; Verheecke, C.; Sabaou, N.; Mathieu, F. Contamination of common spices by aflatoxigenic fungi and aflatoxin B1 in Algeria. Qual. Assur. Saf. Crop. Foods 2016, 8, 137–144. [Google Scholar] [CrossRef] [Green Version]
- Yu, J. Current understanding on aflatoxin biosynthesis and future perspective in reducing aflatoxin contamination. Toxins 2012, 4, 1024–1057. [Google Scholar] [CrossRef] [Green Version]
- Agriopoulou, S.; Stamatelopoulou, E.; Varzakas, T. Advances in occurrence, importance, and mycotoxin control strategies: Prevention and detoxification in foods. Foods 2020, 9, 137. [Google Scholar] [CrossRef]
- European Commission. RASFF—Food and Feed Safety Alerts|Food Safety. 2016. Available online: https://ec.europa.eu/food/safety/rasff_en (accessed on 14 April 2020).
- Battilani, P.; Toscano, P.; Van Der Fels-Klerx, H.J.; Moretti, A.; Leggieri, M.C.; Brera, C.; Rortais, A.; Goumperis, T.; Robinson, T. Aflatoxin B 1 contamination in maize in Europe increases due to climate change. Sci. Rep. 2016, 6, 24328. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- European Commission. Maximum Levels for Certain Contaminants in Foodsuffs. Available online: https://eur-lex.europa.eu/legal-content/EN/ALL/?uri=celex:32006R1881 (accessed on 16 April 2020).
- Birck, N.M.M.; Lorini, I.; Scussel, V.M. Fungus and mycotoxins in wheat grain at post harvest. In Proceedings of the 9th International Working Conference on Stored Product Protection, Campinas, Brazil, 15–18 October 2006; pp. 198–205. [Google Scholar]
- Ren, X.; Zhang, Q.; Zhang, W.; Mao, J.; Li, P. Control of aflatoxigenic molds by antagonistic microorganisms: Inhibitory behaviors, bioactive compounds, related mechanisms, and influencing factors. Toxins 2020, 12, 24. [Google Scholar] [CrossRef] [Green Version]
- Wu, Q.; Jezkova, A.; Yuan, Z.; Pavlikova, L.; Dohnal, V.; Kuca, K. Biological degradation of aflatoxins. Drug Metab. Rev. 2009, 41, 1–7. [Google Scholar] [CrossRef]
- Eshelli, M.; Harvey, L.; Edrada-Ebel, R.; McNeil, B. Metabolomics of the bio-degradation process of aflatoxin B1 by actinomycetes at an initial pH of 6.0. Toxins 2015, 7, 439–456. [Google Scholar] [CrossRef] [Green Version]
- Verheecke, C.; Liboz, T.; Mathieu, F. Microbial degradation of aflatoxin B1: Current status and future advances. Int. J. Food Microbiol. 2016, 237, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Barka, E.A.; Vatsa, P.; Sanchez, L.; Gaveau-Vaillant, N.; Jacquard, C.; Klenk, H.-P.; Clément, C.; Ouhdouch, Y.; Van Wezel, G.P. Taxonomy, Physiology, and Natural Products of Actinobacteria. Microbiol. Mol. Biol. Rev. 2016, 80, 1–43. [Google Scholar] [CrossRef] [Green Version]
- Verheecke, C.; Liboz, T.; Anson, P.; Zhu, Y.; Mathieu, F. Streptomyces–Aspergillus flavus interactions: Impact on aflatoxin B accumulation. Food Addit. Contam. Part A 2015, 32, 572–576. [Google Scholar] [CrossRef]
- Verheecke, C.; Liboz, T.; Darriet, M.; Sabaou, N.; Mathieu, F. In vitro interaction of actinomycetes isolates with Aspergillus flavus: Impact on aflatoxins B1 and B2 production. Lett. Appl. Microbiol. 2014, 58, 597–603. [Google Scholar] [CrossRef] [Green Version]
- Wongsariya, K.; Thawai, C. Antifungal Activity against the Growth of Aflatoxin Producing Fungi from Soil Actinobacteria. J. Adv. Agric. Technol. 2019, 6, 200–204. [Google Scholar] [CrossRef]
- Sakuda, S.; Ono, M.; Furihata, K.; Nakayama, J.; Suzuki, A.; Isogai, A. Aflastatin A, a novel inhibitor of aflatoxin production of Aspergillus parasiticus, from Streptomyces. J. Am. Chem. Soc. 1996, 118, 7855–7856. [Google Scholar] [CrossRef]
- Sakuda, S. Mycotoxin production inhbitors from natural products. Mycotoxins 2010, 60, 79–86. [Google Scholar] [CrossRef] [Green Version]
- Caceres, I.; Snini, S.P.; Puel, O.; Mathieu, F. Streptomyces roseolus, A Promising Biocontrol Agent against Aspergillus flavus, the Main Aflatoxin B1 Producer. Toxins 2018, 10, 442. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vanhoutte, I.; Audenaert, K.; De Gelder, L. Biodegradation of mycotoxins: Tales from known and unexplored worlds. Front. Microbiol. 2016, 7. [Google Scholar] [CrossRef] [Green Version]
- Adebo, O.A.; Njobeh, P.B.; Gbashi, S.; Nwinyi, O.C.; Mavumengwana, V. Review on microbial degradation of aflatoxins. Crit. Rev. Food Sci. Nutr. 2017, 57, 3208–3217. [Google Scholar] [CrossRef] [PubMed]
- Vincenzi, A.; Silva, F.; Naira, L.; Oliveira, C.A.F. Biomarkers of Aflatoxin Exposure and Its Relationship with the Hepatocellular Carcinoma. In Aflatoxins—Biochemistry and Molecular Biology; InTech: Rijeka, Croatia, 2011. [Google Scholar]
- Krifaton, C.; Kriszt, B.; Szoboszlay, S.; Cserháti, M.; Szűcs, Á.; Kukolya, J. Analysis of aflatoxin-B1-degrading microbes by use of a combined toxicity-profiling method. Mutat. Res. Genet. Toxicol. Environ. Mutagen. 2011, 726, 1–7. [Google Scholar] [CrossRef]
- Quillardet, P.; De Bellecombe, C.; Hofnung, M. The SOS Chromotest, a colorimetric bacterial assay for genotoxins: Validation study with 83 compounds. Mutat. Res. Mutagen. Relat. Subj. 1985, 147, 79–95. [Google Scholar] [CrossRef]
- Quillardet, P.; Hofnung, M. The SOS chromotest: A review. Mutat. Res. Rev. Genet. Toxicol. 1993, 297, 235–279. [Google Scholar] [CrossRef]
- Campos-Avelar, I.; De La Noue, A.C.; Durand, N.; Fay, B.; Martinez, V.; Fontana, A.; Strub, C.; Schorr-Galindo, S. Minimizing ochratoxin a contamination through the use of actinobacteria and their active molecules. Toxins 2020, 12, 296. [Google Scholar] [CrossRef]
- Manteca, A.; Yague, P. Streptomyces differentiation in liquid cultures as a trigger of secondary metabolism. Antibiotics 2018, 7, 41. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wakefield, J.; Hassan, H.M.; Jaspars, M.; Ebel, R.; Rateb, M.E. Dual induction of new microbial secondary metabolites by fungal bacterial co-cultivation. Front. Microbiol. 2017, 8, 1284. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nazari, B.; Saito, A.; Kobayashi, M.; Miyashita, K.; Wang, Y.; Fujii, T. High expression levels of chitinase genes in Streptomyces coelicolor A3(2) grown in soil. FEMS Microbiol. Ecol. 2011, 77, 623–635. [Google Scholar] [CrossRef]
- Williamson, N.; Brian, P.; Wellington, E. Molecular detection of bacterial and streptomycete chitinases in the environment. Antonie Van Leeuwenhoek 2000, 78, 315–321. [Google Scholar] [CrossRef]
- Prapagdee, B.; Kuekulvong, C.; Mongkolsuk, S. Antifungal potential of extracellular metabolites produced by Streptomyces hygroscopicus against phytopathogenic fungi. Int. J. Biol. Sci. 2008, 4, 330–337. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boukaew, S.; Prasertsan, P. Suppression of rice sheath blight disease using a heat stable culture filtrate from Streptomyces philanthi RM-1-138. Crop Prot. 2014, 61, 1–10. [Google Scholar] [CrossRef]
- Sangare, L.; Zhao, Y.; Folly, Y.M.E.; Chang, J.; Li, J.; Selvaraj, J.N.; Xing, F.; Zhou, L.; Wang, Y.; Liu, Y. Aflatoxin B₁ degradation by a Pseudomonas strain. Toxins 2014, 6, 3028–3040. [Google Scholar] [CrossRef] [PubMed]
- Wheeler, K.A.; Hurdman, B.F.; Pitt, J. Influence of pH on the growth of some toxigenic species of Aspergillus, Penicillium and Fusarium. Int. J. Food Microbiol. 1991, 12, 141–149. [Google Scholar] [CrossRef]
- Nguyen, P.-A.; Strub, C.; Fontana, A.; Schorr-Galindo, S. Crop molds and mycotoxins: Alternative management using biocontrol. Biol. Control 2017, 104, 10–27. [Google Scholar] [CrossRef]
- Olivier, P.; Marzin, D. Study of the genotoxic potential of 48 inorganic derivatives with the SOS chromotest. Mutat. Res. Toxicol. 1987, 189, 263–269. [Google Scholar] [CrossRef]
- Davis, N.D.; Diener, U.L.; Agnihotri, V.P. Production of aflatoxins B1 and G1 in chemically defined medium. Mycopathol. Mycol. Appl. 1967, 31, 251–256. [Google Scholar] [CrossRef] [PubMed]
- Yoshinari, T.; Noda, Y.; Yoda, K.; Sezaki, H.; Nagasawa, H.; Sakuda, S. Inhibitory activity of blasticidin A, a strong aflatoxin production inhibitor, on protein synthesis of yeast: Selective inhibition of aflatoxin production by protein synthesis inhibitors. J. Antibiot. 2010, 63, 309–314. [Google Scholar] [CrossRef] [PubMed]
- Yoshinari, T.; Akiyama, T.; Nakamura, K.; Kondo, T.; Takahashi, Y.; Muraoka, Y.; Nonomura, Y.; Nagasawa, H.; Sakuda, S. Dioctatin A is a strong inhibitor of aflatoxin production by Aspergillus parasiticus. Microbiology 2007, 153, 2774–2780. [Google Scholar] [CrossRef] [Green Version]
- Woloshuk, C.P.; Shim, W.B. Aflatoxins, fumonisins, and trichothecenes: A convergence of knowledge. FEMS Microbiol. Rev. 2013, 37, 94–109. [Google Scholar] [CrossRef] [Green Version]
- Keller, N.P.; Nesbitt, C.; Sarr, B.; Phillips, T.D.; Burow, G.B. pH regulation of sterigmatocystin and aflatoxin biosynthesis in Aspergillus spp. Phytopathology 1997, 87, 643–648. [Google Scholar] [CrossRef] [Green Version]
- Pfliegler, W.P.; Pócsi, I.; Győri, Z.; Pusztahelyi, T. The Aspergilli and Their Mycotoxins: Metabolic Interactions With Plants and the Soil Biota. Front. Microbiol. 2020, 10. [Google Scholar] [CrossRef] [Green Version]
- Van Rij, E.T.; Girard, G.; Lugtenberg, B.J.J.; Bloemberg, G.V. Influence of fusaric acid on phenazine-1-carboxamide synthesis and gene expression of Pseudomonas chlororaphis strain PCL1391. Microbiology 2005, 151, 2805–2814. [Google Scholar] [CrossRef] [Green Version]
- Rasmussen, T.B.; Skindersoe, M.E.; Bjarnsholt, T.; Phipps, R.K.; Christensen, K.B.; Jensen, P.O.; Andersen, J.B.; Koch, B.; Larsen, T.O.; Hentzer, M.; et al. Identity and effects of quorum-sensing inhibitors produced by Penicillium species. Microbiology 2005, 151, 1325–1340. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sweany, R.R.; Damann, K.E. Influence of Neighboring Clonal-Colonies on Aflatoxin Production by Aspergillus flavus. Front. Microbiol. 2020, 10, 3038. [Google Scholar] [CrossRef] [Green Version]
- Medina, A.; Mohale, S.; Samsudin, N.I.P.; Rodriguez-Sixtos, A.; Rodriguez, A.; Magan, N. Biocontrol of mycotoxins: Dynamics and mechanisms of action. Curr. Opin. Food Sci. 2017, 17, 41–48. [Google Scholar] [CrossRef]
- Al-Saad, L.A.; Al-Badran, A.I.; Al-Jumayli, S.A.; Magan, N.; Rodríguez, A. Impact of bacterial biocontrol agents on aflatoxin biosynthetic genes, aflD and aflR expression, and phenotypic aflatoxin B1 production by Aspergillus flavus under different environmental and nutritional regimes. Int. J. Food Microbiol. 2016, 217, 123–129. [Google Scholar] [CrossRef] [PubMed]
- Harkai, P.; Szabó, I.; Cserháti, M.; Krifaton, C.; Risa, A.; Radó, J.; Balázs, A.; Berta, K.; Kriszt, B. Biodegradation of aflatoxin-B1 and zearalenone by Streptomyces sp. collection. Int. Biodeterior. Biodegrad. 2016, 108, 48–56. [Google Scholar] [CrossRef] [Green Version]
- Teniola, O.D.; Addo, P.A.; Brost, I.M.; Farber, P.; Jany, K.-D.; Alberts, J.F.; Van Zyl, W.H.; Steyn, P.S. Degradation of aflatoxin B1 by cell-free extracts of Rhodococcus erythropolis and Mycobacterium fluoranthenivorans sp. nov. DSM44556T. Int. J. Food Microbiol. 2005, 105, 111–117. [Google Scholar] [CrossRef]
- El Khoury, R.; Mathieu, F.; Atoui, A.; Kawtharani, H.; El Khoury, A.; Afif, C.; Maroun, R.G.; El Khoury, A. Ability of soil isolated actinobacterial strains to prevent, bind and biodegrade ochratoxin A. Toxins 2017, 9, 222. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Manteca, A.; Alvarez, R.; Salazar, N.; Yagüe, P.; Sanchez, J. Mycelium differentiation and antibiotic production in submerged cultures of Streptomyces coelicolor. Appl. Environ. Microbiol. 2008, 74, 3877–3886. [Google Scholar] [CrossRef] [Green Version]
- Mannaa, M.; Kim, K.D. Influence of temperature and water activity on deleterious fungi and mycotoxin production during grain storage. Mycobiology 2017, 45, 240–254. [Google Scholar] [CrossRef] [PubMed]
- Alberts, J.F.; Engelbrecht, Y.; Steyn, P.S.; Holzapfel, W.H.; Van Zyl, W.H. Biological degradation of aflatoxin B1 by Rhodococcus erythropolis cultures. Int. J. Food Microbiol. 2006, 109, 121–126. [Google Scholar] [CrossRef]
- Taylor, M.C.; Jackson, C.J.; Tattersall, D.B.; French, N.; Peat, T.S.; Newman, J.; Briggs, L.J.; Lapalikar, G.V.; Campbell, P.M.; Scott, C.; et al. Identification and characterization of two families of F420H2-dependent reductases from Mycobacteria that catalyse aflatoxin degradation. Mol. Microbiol. 2010, 78, 561–575. [Google Scholar] [CrossRef] [Green Version]
- Lapalikar, G.V.; Taylor, M.C.; Warden, A.C.; Scott, C.; Russell, R.J.; Oakeshott, J.G. F 420H 2-dependent degradation of aflatoxin and other furanocoumarins is widespread throughout the Actinomycetales. PLoS ONE 2012, 7. [Google Scholar] [CrossRef] [Green Version]
- Guan, S.; Ji, C.; Zhou, T.; Li, J.; Ma, Q.; Niu, T. Aflatoxin B 1 degradation by Stenotrophomonas maltophilia and other microbes selected using coumarin medium. Int. J. Mol. Sci. 2008, 9, 1489–1503. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, L.; Wu, J.; Liu, Z.; Shi, Y.; Liu, J.; Xu, X.; Hao, S.; Mu, P.; Deng, F.; Deng, Y. Aflatoxin B1 degradation and detoxification by Escherichia coli CG1061 isolated from chicken cecum. Front. Pharmacol. 2019, 9, 1548. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Hassan, Y.I.; Lepp, D.; Shao, S.; Zhou, T. Strategies and methodologies for developing microbial detoxification systems to mitigate mycotoxins. Toxins 2017, 9, 130. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, L.; Guan, S.; Gao, X.; Ma, Q.; Lei, Y.; Bai, X.; Ji, C. Preparation, purification and characteristics of an aflatoxin degradation enzyme from Myxococcus fulvus ANSM068. J. Appl. Microbiol. 2011, 110, 147–155. [Google Scholar] [CrossRef]
- Loi, M.; Fanelli, F.; Liuzzi, V.C.; Logrieco, A.F.; Mulè, G. Mycotoxin biotransformation by native and commercial enzymes: Present and future perspectives. Toxins 2017, 9, 111. [Google Scholar] [CrossRef]
- Samuel, M.S.; Sivaramakrishna, A.; Mehta, A. Degradation and detoxification of aflatoxin B1 by Pseudomonas putida. Int. Biodeterior. Biodegrad. 2014, 86, 202–209. [Google Scholar] [CrossRef]
- Lee, L.S.; Dunn, J.J.; Delucca, A.J.; Ciegler, A. Role of lactone ring of aflatoxin B1 in toxicity and mutagenicity. Experientia 1981, 37, 16–17. [Google Scholar] [CrossRef] [PubMed]
- Iram, W.; Anjum, T.; Iqbal, M.; Ghaffar, A.; Abbas, M. Mass spectrometric identification and toxicity assessment of degraded products of aflatoxin B1 and B2 by Corymbia citriodora aqueous extracts. Sci. Rep. 2015, 5, 14672. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iram, W.; Anjum, T.; Iqbal, M.; Ghaffar, A.; Abbas, M.; Khan, A.M. Structural analysis and biological toxicity of aflatoxins B1 and B2 degradation products following detoxification by Ocimum basilicum and Cassia fistula aqueous extracts. Front. Microbiol. 2016, 7. [Google Scholar] [CrossRef]
- Iram, W.; Anjum, T.; Iqbal, M.; Ghaffar, A.; Abbas, M. Structural elucidation and toxicity assessment of degraded products of aflatoxin B1 and B2 by aqueous extracts of Trachyspermum ammi. Front. Microbiol. 2016, 7, 346. [Google Scholar] [CrossRef] [Green Version]
- Wall-Martínez, H.; Ramírez-Martínez, A.; Wesolek, N.; Brabet, C.; Durand, N.; Rodríguez-Jimenes, G.C.; García-Alvarado, M.A.; Salgado-Cervantes, M.A.; Robles-Olvera, V.J.; Roudot, A. Risk assessment of exposure to mycotoxins (aflatoxins and fumonisins) through corn tortilla intake in Veracruz City (Mexico). Food Addit. Contam. Part A 2019, 36, 929–939. [Google Scholar] [CrossRef]









| Cluster | Specific Features | Subcluster | Specific Features | Isolate/Strain | |
|---|---|---|---|---|---|
| A | 1 | Limited inhibition of A. flavus growth. Moderate decrease in AFB1sp in dual culture. Limited degradation capacity. | IX16, IX37, IX38, IX47, IX53, IX57 | ||
| I | Moderate effect on A. flavus growth. Contrasted effect on AFB1-specific production and its degradation. | 2 | Lack of inhibition of A. flavus growth. Strong decrease in AFB1sp in dual culture. Moderate capacity of CFEs to decrease AFB1sp. Moderate degradation capacity of bacterial cells. | IX30, IX40, IX41, IX43, IX44 | |
| B | 1 | Moderate decrease in AFB1sp in dual culture. Strong increase in AFB1sp provoked by CFEs. Strong degradation capacity of bacterial cells. | IX05, IX54, MYC | ||
| 2 | Moderate decrease in AFB1sp. Strong degradation capacity of bacterial cells. | IX01, IX03, IX04, IX12, IX20, IX25, IX28, IX35, IX36, IX39, IX45, IX58 | |||
| 3 | Strong decrease in AFB1sp in dual culture and by unheated CFEs. Moderate degradation capacities. | IX02, IX06, IX08, IX29, IX33, IX34, IX42, IX46 | |||
| II | Strong inhibition of A. flavus growth (dual culture). Strong effect on AFB1sp. Moderate degradation by bacterial cells and CFEs. | A | Strong inhibition of A. flavus growth in dual culture. Strong decrease in AFB1sp in dual culture. Moderate degradation by bacterial cells and by CFEs. | IX14, IX22, IX23, IX48, IX50, IX55, IX56, IX59 | |
| B | Strong inhibition of A. flavus growth in dual culture. Strong decrease in AFB1sp in dual culture. Moderate decrease in AFB1sp by CFEs. Moderate degradation by bacterial cells and by CFEs. | IX07, IX09, IX10, IX11, IX13, IX15, IX17, IX18, IX19, IX21, IX24, IX26, IX27, IX31, IX32, IX49, IX51, IX52 | |||
| Streptomyces Isolate/Strain Reduction in Genotoxicity | |||||
|---|---|---|---|---|---|
| Standard AFB1 | Control | IX20 | IX45 | MYC | |
| Residual AFB1 (µg/mL) | 8 | 7.71 ± 0.22 | 0.98 ± 0.09 | 0.32 ± 0.04 | 1.95 ± 1.65 |
| IF | 7.13 | 6.05 ± 0.27 | 2.26 ± 0.60 | 1.99 ± 0.53 | 2.18 ± 0.11 |
| % IF | 100 | 85 ± 4 | 32 ± 8 | 28 ± 7 | 31 ± 2 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Campos-Avelar, I.; Colas de la Noue, A.; Durand, N.; Cazals, G.; Martinez, V.; Strub, C.; Fontana, A.; Schorr-Galindo, S. Aspergillus flavus Growth Inhibition and Aflatoxin B1 Decontamination by Streptomyces Isolates and Their Metabolites. Toxins 2021, 13, 340. https://doi.org/10.3390/toxins13050340
Campos-Avelar I, Colas de la Noue A, Durand N, Cazals G, Martinez V, Strub C, Fontana A, Schorr-Galindo S. Aspergillus flavus Growth Inhibition and Aflatoxin B1 Decontamination by Streptomyces Isolates and Their Metabolites. Toxins. 2021; 13(5):340. https://doi.org/10.3390/toxins13050340
Chicago/Turabian StyleCampos-Avelar, Ixchel, Alexandre Colas de la Noue, Noël Durand, Guillaume Cazals, Véronique Martinez, Caroline Strub, Angélique Fontana, and Sabine Schorr-Galindo. 2021. "Aspergillus flavus Growth Inhibition and Aflatoxin B1 Decontamination by Streptomyces Isolates and Their Metabolites" Toxins 13, no. 5: 340. https://doi.org/10.3390/toxins13050340
APA StyleCampos-Avelar, I., Colas de la Noue, A., Durand, N., Cazals, G., Martinez, V., Strub, C., Fontana, A., & Schorr-Galindo, S. (2021). Aspergillus flavus Growth Inhibition and Aflatoxin B1 Decontamination by Streptomyces Isolates and Their Metabolites. Toxins, 13(5), 340. https://doi.org/10.3390/toxins13050340