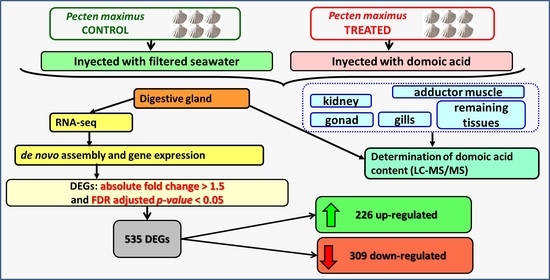
Transcriptional Response in the Digestive Gland of the King Scallop (Pecten maximus) After the Injection of Domoic Acid
Abstract
:1. Introduction
2. Results
2.1. Domoic Acid Content in the Tissues of P. maximus
2.2. Sequencing and De Novo Assembly
2.3. Differential Expression and Functional Annotation
2.4. Protein Network Analysis
2.5. Real Time RT-qPCR
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Animals
5.2. Determination of the Domoic Acid Content
5.3. RNA Extraction
5.4. Library Preparation and Sequencing
5.5. De Novo Assembly
5.6. Differential Expression
5.7. Functional Annotation
5.8. Protein Network Analysis
5.9. Technical Validation of RNA-seq data by RT-qPCR
5.10. Statistical Analyses
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bates, S.S.; Bird, C.J.; de Freitas, A.S.W.; Foxall, R.; Gilgan, M.; Hanic, L.A.; Johnson, G.R.; McCulloch, A.W.; Odense, P.; Pocklington, R.; et al. Pennate Diatom Nitzschia pungens as the Primary Source of Domoic Acid, a Toxin in Shellfish from Eastern Prince Edward Island, Canada. Can. J. Fish. Aquat. Sci. 1989, 46, 1203–1215. [Google Scholar] [CrossRef]
- Bates, S.S.; Hubbard, K.A.; Lundholm, N.; Montresor, M.; Leaw, C.P. Pseudo-Nitzschia, Nitzschia, and domoic acid: New research since 2011. Harmful Algae 2018, 79, 3–43. [Google Scholar] [CrossRef] [PubMed]
- Lelong, A.; Hégaret, H.; Soudant, P.; Bates, S.S. Pseudo-Nitzschia (Bacillariophyceae) species, domoic acid and amnesic shellfish poisoning: Revisiting previous paradigms. Phycologia 2012, 51, 168–216. [Google Scholar] [CrossRef] [Green Version]
- Pulido, O.M. Domoic Acid Toxicologic Pathology: A Review. Mar. Drugs 2008, 6, 180–219. [Google Scholar] [CrossRef]
- Trainer, V.L.; Bates, S.S.; Lundholm, N.; Thessen, A.E.; Cochlan, W.P.; Adams, N.G.; Trick, C.G. Pseudo-Nitzschia physiological ecology, phylogeny, toxicity, monitoring and impacts on ecosystem health. Harmful Algae 2012, 14, 271–300. [Google Scholar] [CrossRef] [Green Version]
- Zabaglo, K.; Chrapusta, E.; Bober, B.; Kaminski, A.; Adamski, M.; Bialczyk, J. Environmental roles and biological activity of domoic acid: A review. Algal Res. 2016, 13, 94–101. [Google Scholar] [CrossRef]
- Costa, L.G.; Giordano, G.; Faustman, E.M. Domoic acid as a developmental neurotoxin. NeuroToxicology 2010, 31, 409–423. [Google Scholar] [CrossRef] [Green Version]
- Lefebvre, K.A.; Robertson, A. Domoic acid and human exposure risks: A review. Toxicon 2010, 56, 218–230. [Google Scholar] [CrossRef]
- Duncan, P.F.; Brand, A.R.; Strand, Ø.; Foucher, E. Chapter 19—The European Scallop Fisheries for Pecten maximus, Aequipecten opercularis, Chlamys islandica, and Mimachlamys varia. In Developments in Aquaculture and Fisheries Science; Shumway, S.E., Parsons, G.J., Eds.; Elsevier: Amsterdam, The Netherlands, 2016; Volume 40, pp. 781–858. [Google Scholar]
- Novaczek, I.; Madhyastha, M.; Ablett, R.; Johnson, G.; Nijjar, M.; Sims, D. Uptake, disposition and depuration of domoic acid by blue mussels (Mytilus edulis). Aquat. Toxicol. 1991, 21, 103–118. [Google Scholar] [CrossRef]
- Novaczek, I.; Madhyastha, M.S.; Ablett, R.F.; Donald, A.; Johnson, G.; Nijjar, M.S.; Sims, D.E. Depuration of Domoic Acid from Live Blue Mussels (Mytilus edulis). Can. J. Fish. Aquat. Sci. 1992, 49, 312–318. [Google Scholar] [CrossRef]
- Blanco, J.; de la Puente, M.B.; Arévalo, F.; Salgado, C.; Moroño, Á. Depuration of Mussels (Mytilus galloprovincialis) Con-taminated with Domoic Acid. Aquat. Living Resour. 2002, 15, 53–60. [Google Scholar] [CrossRef]
- Mafra, L.L., Jr.; Bricelj, V.M.; Fennel, K. Domoic acid uptake and elimination kinetics in oysters and mussels in relation to body size and anatomical distribution of toxin. Aquat. Toxicol. 2010, 100, 17–29. [Google Scholar] [CrossRef] [Green Version]
- Blanco, J.; Acosta, C.P.; Bermúdez de la Puente, M.; Salgado, C. Depuration and Anatomical Distribution of the Amnesic Shellfish Poisoning (ASP) Toxin Domoic Acid in the King Scallop Pecten Maximus. Aquat. Toxicol. 2002, 60, 111–121. [Google Scholar] [CrossRef]
- Blanco, J.; Acosta, C.P.; Mariño, C.; Muñiz, S.; Martín, H.; Moroño, Á.; Correa, J.; Arévalo, F.; Salgado, C. Depuration of domoic acid from different body compartments of the king scallop Pecten maximusgrown in raft culture and natural bed. Aquat. Living Resour. 2006, 19, 257–265. [Google Scholar] [CrossRef]
- Blanco, J.; Mauríz, A.; Álvarez, G. Distribution of Domoic Acid in the Digestive Gland of the King Scallop Pecten maximus. Toxins 2020, 12, 371. [Google Scholar] [CrossRef]
- Álvarez, G.; Rengel, J.; Araya, M.; Álvarez, F.; Pino, R.; Uribe, E.; Díaz, P.A.; Rossignoli, A.E.; López-Rivera, A.; Blanco, J. Rapid Domoic Acid Depuration in the Scallop Argopecten purpuratus and Its Transfer from the Digestive Gland to Other Organs. Toxins 2020, 12, 698. [Google Scholar] [CrossRef]
- Douglas, D.J.; Kenchington, E.R.; Bird, C.J.; Pocklington, R.; Bradford, B.; Silvert, W. Accumulation of domoic acid by the sea scallop (Placopecten magellanicus) fed cultured cells of toxic Pseudo-Nitzschia multiseries. Can. J. Fish. Aquat. Sci. 1997, 54, 907–913. [Google Scholar] [CrossRef]
- Madhyastha, M.; Novaczek, I.; Ablett, R.; Johnson, G.; Nijjar, M.; Sims, D. In vitro study of domoic acid uptake by gland tissue of blue mussel (Mytilus L.). Aquat. Toxicol. 1991, 20, 73–81. [Google Scholar] [CrossRef]
- Wright, J.L.C.; Boyd, R.K.; de Freitas, A.S.W.; Falk, M.; Foxall, R.A.; Jamieson, W.D.; Laycock, M.V.; McCulloch, A.W.; McInnes, A.G.; Odense, P.; et al. Identification of domoic acid, a neuroexcitatory amino acid, in toxic mussels from eastern Prince Edward Island. Can. J. Chem. 1989, 67, 481–490. [Google Scholar] [CrossRef]
- Mauriz, A.; Blanco, J. Distribution and linkage of domoic acid (amnesic shellfish poisoning toxins) in subcellular fractions of the digestive gland of the scallop Pecten maximus. Toxicon 2010, 55, 606–611. [Google Scholar] [CrossRef]
- Dizer, H.; Fischer, B.; Harabawy, A.; Hennion, M.-C.; Hansen, P.-D. Toxicity of domoic acid in the marine mussel Mytilus edulis. Aquat. Toxicol. 2001, 55, 149–156. [Google Scholar] [CrossRef]
- Jones, T.; Whyte, J.; Townsend, L.; Ginther, N.; Iwama, G. Effects of domoic acid on haemolymph pH, PCO2 and PO2 in the Pacific oyster, Crassostrea gigas and the California mussel, Mytilus californianus. Aquat. Toxicol. 1995, 31, 43–55. [Google Scholar] [CrossRef]
- Jones, T.O.; Whyte, J.N.; Ginther, N.G.; Townsend, L.D.; Iwama, G.K. Haemocyte changes in the pacific oyster, crassostrea gigas, caused by exposure to domoic acid in the diatom Pseudo-Nitzschia pungens f. multiseries. Toxicon 1995, 33, 347–353. [Google Scholar] [CrossRef]
- Liu, H.; Kelly, M.S.; Campbell, D.A.; Fang, J.; Zhu, J. Accumulation of Domoic Acid and Its Effect on Juvenile King Scallop<I> Pecten Maximus (Linnaeus, 1758). Aquaculture 2008, 284, 224–230. [Google Scholar]
- Liu, H.; Kelly, M.S.; Campbell, D.A.; Dong, S.L.; Zhu, J.X.; Wang, S.F. Exposure to domoic acid affects larval development of king scallop Pecten maximus (Linnaeus, 1758). Aquat. Toxicol. 2007, 81, 152–158. [Google Scholar] [CrossRef]
- Chi, C.; Zhang, C.; Liu, J.; Zheng, X. Effects of Marine Toxin Domoic Acid on Innate Immune Responses in Bay Scallop Argopecten irradians. J. Mar. Sci. Eng. 2019, 7, 407. [Google Scholar] [CrossRef] [Green Version]
- Song, J.A.; Choi, C.Y.; Park, H.-S. Exposure to domoic acid causes oxidative stress in bay scallops Argopecten irradians. Fish. Sci. 2020, 86, 701–709. [Google Scholar] [CrossRef]
- Pazos, A.J.; Ventoso, P.; Martínez-Escauriaza, R.; Pérez-Parallé, M.L.; Blanco, J.; Triviño, J.C.; Sánchez, J.L. Transcriptional response after exposure to domoic acid-producing Pseudo-Nitzschia in the digestive gland of the mussel Mytilus galloprovincialis. Toxicon 2017, 140, 60–71. [Google Scholar] [CrossRef]
- Ventoso, P.; Pazos, A.J.; Pérez-Parallé, M.L.; Blanco, J.; Triviño, J.C.; Sánchez, J.L. RNA-Seq Transcriptome Profiling of the Queen Scallop (Aequipecten Opercularis) Digestive Gland after Exposure to Domoic Acid-Producing Pseudo-Nitzschia. Toxins 2019, 11, 97. [Google Scholar] [CrossRef] [Green Version]
- Hiolski, E.M.; Kendrick, P.S.; Frame, E.R.; Myers, M.S.; Bammler, T.K.; Beyer, R.P.; Farin, F.M.; Wilkerson, H.-W.; Smith, D.R.; Marcinek, D.J.; et al. Chronic low-level domoic acid exposure alters gene transcription and impairs mitochondrial function in the CNS. Aquat. Toxicol. 2014, 155, 151–159. [Google Scholar] [CrossRef] [Green Version]
- Giordano, G.; White, C.C.; Mohar, I.; Kavanagh, T.J.; Costa, L.G. Glutathione Levels Modulate Domoic Acid–Induced Apoptosis in Mouse Cerebellar Granule Cells. Toxicol. Sci. 2007, 100, 433–444. [Google Scholar] [CrossRef]
- Giordano, G.; White, C.C.; McConnachie, L.A.; Fernandez, C.; Kavanagh, T.J.; Costa, L.G. Neurotoxicity of Domoic Acid in Cerebellar Granule Neurons in a Genetic Model of Glutathione Deficiency. Mol. Pharmacol. 2006, 70, 2116–2126. [Google Scholar] [CrossRef] [Green Version]
- Xu, R.; Tao, Y.; Wu, C.; Yi, J.; Yang, Y.; Yang, R.; Hong, D. Domoic acid induced spinal cord lesions in adult mice: Evidence for the possible molecular pathways of excitatory amino acids in spinal cord lesions. NeuroToxicology 2008, 29, 700–707. [Google Scholar] [CrossRef]
- Cabrera, J.; González, P.M.; Puntarulo, S. The Phycotoxin Domoic Acid as a Potential Factor for Oxidative Alterations Enhanced by Climate Change. Front. Plant. Sci. 2020, 11, 576971. [Google Scholar] [CrossRef]
- Tian, D.; Zhang, G. Toxic Effects of Domoic Acid on Caenorhabditis elegans and the Underlying Mechanism. Int. J. Biol. 2019, 11, v11n3p1. [Google Scholar] [CrossRef]
- Pramod, A.B.; Foster, J.; Carvelli, L.; Henry, L.K. SLC6 transporters: Structure, function, regulation, disease association and therapeutics. Mol. Asp. Med. 2013, 34, 197–219. [Google Scholar] [CrossRef] [Green Version]
- Franceschini, A.; Szklarczyk, D.; Frankild, S.; Kuhn, M.; Simonovic, M.; Roth, A.; Lin, J.; Minguez, P.; Bork, P.; von Mering, C.; et al. STRING v9.1: Protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 2012, 41, D808–D815. [Google Scholar] [CrossRef] [Green Version]
- Mauriz, O.; Maneiro, V.; Pérez-Parallé, M.L.; Sanchez, J.L.; Pazos, A.J. Selection of reference genes for quantitative RT-PCR studies on the gonad of the bivalve mollusc Pecten maximus L. Aquaculture 2012, 370-371, 158–165. [Google Scholar] [CrossRef]
- Beninger, P.G.; Le Pennec, M. Chapter 3—Scallop Structure and Function. In Developments in Aquaculture and Fisheries Science; Shumway, S.E., Parsons, G.J., Eds.; Elsevier: Amsterdam, The Netherlands, 2016; Volume 40, pp. 85–159. [Google Scholar]
- Lefebvre, K.A.; Tilton, S.C.; Bammler, T.K.; Beyer, R.P.; Srinouanprachan, S.; Stapleton, P.L.; Farin, F.M.; Gallagher, E.P. Gene Expression Profiles in Zebrafish Brain after Acute Exposure to Domoic Acid at Symptomatic and Asymptomatic Doses. Toxicol. Sci. 2008, 107, 65–77. [Google Scholar] [CrossRef]
- Pérez-Gómez, A.; Tasker, R.A. Domoic Acid as a Neurotoxin. In Handbook of Neurotoxicity; Kostrzewa, R.M., Ed.; Springer: New York, NY, USA, 2014; pp. 399–419. [Google Scholar]
- Jeffery, B.; Barlow, T.; Moizer, K.; Paul, S.; Boyle, C. Amnesic shellfish poison. Food Chem. Toxicol. 2004, 42, 545–557. [Google Scholar] [CrossRef]
- Ryan, J.; Morey, J.; Ramsdell, J.; van Dolah, F. Acute phase gene expression in mice exposed to the marine neurotoxin domoic acid. Neuroscience 2005, 136, 1121–1132. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Huang, Y.; Liang, X.-F.; Li, S.-Y.; Ip, K.-C. Transcriptional responses of xenobiotic metabolizing enzymes, HSP70 and Na+/K+-ATPase in the liver of rabbitfish (Siganus oramin) intracoelomically injected with amnesic shellfish poisoning toxin. Environ. Toxicol. 2008, 23, 363–371. [Google Scholar] [CrossRef] [PubMed]
- Anderson, K.; Taylor, D.A.; Thompson, E.L.; Melwani, A.R.; Nair, S.V.; Raftos, D.A. Meta-Analysis of Studies Using Suppression Subtractive Hybridization and Microarrays to Investigate the Effects of Environmental Stress on Gene Transcription in Oysters. PLoS ONE 2015, 10, e0118839. [Google Scholar] [CrossRef] [Green Version]
- Tsunekawa, K.; Kondo, F.; Okada, T.; Feng, G.-G.; Huang, L.; Ishikawa, N.; Okada, S. Enhanced expression of WD repeat-containing protein 35 (WDR35) stimulated by domoic acid in rat hippocampus: Involvement of reactive oxygen species generation and p38 mitogen-activated protein kinase activation. BMC Neurosci. 2013, 14, 4. [Google Scholar] [CrossRef] [Green Version]
- Pinto-Silva, C.R.C.; Moukha, S.; Matias, W.G.; Creppy, E.E. Domoic acid induces direct DNA damage and apoptosis in Caco-2 cells: Recent advances. Environ. Toxicol. 2008, 23, 657–663. [Google Scholar] [CrossRef]
- Giordano, G.; Klintworth, H.; Kavanagh, T.; Costa, L. Apoptosis induced by domoic acid in mouse cerebellar granule neurons involves activation of p38 and JNK MAP kinases. Neurochem. Int. 2008, 52, 1100–1105. [Google Scholar] [CrossRef] [Green Version]
- Lein, P.J.; Supasai, S.; Guignet, M. Chapter 9—Apoptosis as a Mechanism of Developmental Neurotoxicity. In Handbook of Developmental Neurotoxicology, 2nd ed.; Slikker, W., Paule, M.G., Wang, C., Eds.; Academic Press: Cambridge, MA, USA, 2018; pp. 91–112. ISBN 978-0-12-809405-1. [Google Scholar]
- Kirchgessner, A.L. Glutamate in the enteric nervous system. Curr. Opin. Pharmacol. 2001, 1, 591–596. [Google Scholar] [CrossRef]
- Storto, M.; Vairetti, M.P.; Sureda, F.X.; Riozzi, B.; Bruno, V.; Nicoletti, F. Expression and Function of Metabotropic Glutamate Receptors in Liver. In Glutamate Receptors in Peripheral Tissue: Excitatory Transmission Outside the CNS; Gill, S., Pulido, O., Eds.; Springer US: Boston, MA, USA, 2007; pp. 211–217. [Google Scholar]
- Du, J.; Li, X.-H.; Li, Y.-J. Glutamate in peripheral organs: Biology and pharmacology. Eur. J. Pharmacol. 2016, 784, 42–48. [Google Scholar] [CrossRef]
- Zafra, F.; Ibáñez, I.; Bartolomé-Martín, D.; Piniella, D.; Arribas-Blázquez, M.; Giménez, C. Glycine Transporters and Its Cou-pling with NMDA Receptors. In Glial Amino Acid Transporters; Advances in Neurobiology; Ortega, A., Schousboe, A., Eds.; Springer International Publishing: Cham, Switzerland, 2017; pp. 55–83. ISBN 978-3-319-55769-4. [Google Scholar]
- Aragón, C.; López-Corcuera, B. Glycine transporters: Crucial roles of pharmacological interest revealed by gene deletion. Trends Pharmacol. Sci. 2005, 26, 283–286. [Google Scholar] [CrossRef]
- Boissonneault, K.R.; Henningsen, B.M.; Bates, S.S.; Robertson, D.L.; Milton, S.; Pelletier, J.; Hogan, D.A.; E Housman, D. Gene expression studies for the analysis of domoic acid production in the marine diatom Pseudo-Nitzschia multiseries. BMC Mol. Biol. 2013, 14, 25. [Google Scholar] [CrossRef] [Green Version]
- Di Dato, V.; Musacchia, F.; Petrosino, G.; Patil, S.; Montresor, M.; Sanges, R.; Ferrante, M.I. Transcriptome sequencing of three Pseudo-Nitzschia species reveals comparable gene sets and the presence of Nitric Oxide Synthase genes in diatoms. Sci. Rep. 2015, 5, 12329. [Google Scholar] [CrossRef]
- Li, Y.; Sun, X.; Zhihui, Y.; Xun, X.; Zhang, J.; Guo, X.; Jiao, W.; Zhang, L.; Liu, W.; Wang, J.; et al. Scallop genome reveals molecular adaptations to semi-sessile life and neurotoxins. Nat. Commun. 2017, 8, 1–11. [Google Scholar] [CrossRef]
- Xun, X.; Cheng, J.; Wang, J.; Li, Y.; Li, X.; Li, M.; Lou, J.; Kong, Y.; Bao, Z.; Hu, X. Solute carriers in scallop genome: Gene expansion and expression regulation after exposure to toxic dinoflagellate. Chemosphere 2020, 241, 124968. [Google Scholar] [CrossRef]
- Vardimon, L.; Ben-Dror, I.; Avisar, N.; Shiftan, L.; Kruchkova, Y.; Oren, A. Regulation of glutamine synthetase in normal and injured neural tissues. Gene Funct. Dis. 2001, 2, 83–88. [Google Scholar] [CrossRef]
- Zou, J.; Wang, Y.-X.; Dou, F.-F.; Lü, H.-Z.; Ma, Z.-W.; Lu, P.-H.; Xu, X.-M. Glutamine synthetase down-regulation reduces astrocyte protection against glutamate excitotoxicity to neurons. Neurochem. Int. 2010, 56, 577–584. [Google Scholar] [CrossRef] [Green Version]
- Fleischer-Lambropoulos, E.; Kazazoglou, T.; Geladopoulos, T.; Kentroti, S.; Stefanis, C.; Vernadakis, A. Stimulation of glutamine synthetase activity by excitatory amino acids in astrocyte cultures derived from aged mouse cerebral hemispheres may be associated with non-n-methyl-d-aspartate receptor activation. Int. J. Dev. Neurosci. 1996, 14, 523–530. [Google Scholar] [CrossRef]
- Lehmann, C.; Bette, S.; Engele, J. High extracellular glutamate modulates expression of glutamate transporters and glutamine synthetase in cultured astrocytes. Brain Res. 2009, 1297, 1–8. [Google Scholar] [CrossRef]
- Hogberg, H.T.; Bal-Price, A.K. Domoic Acid-Induced Neurotoxicity Is Mainly Mediated by the AMPA/KA Receptor: Comparison between Immature and Mature Primary Cultures of Neurons and Glial Cells from Rat Cerebellum. J. Toxicol. 2011, 2011, 543512. [Google Scholar] [CrossRef]
- Krishnan, N.; Dickman, M.B.; Becker, D.F. Proline modulates the intracellular redox environment and protects mammalian cells against oxidative stress. Free Radic. Biol. Med. 2008, 44, 671–681. [Google Scholar] [CrossRef] [Green Version]
- Kenny, N.J.; McCarthy, S.A.; Dudchenko, O.; James, K.; Betteridge, E.; Corton, C.; Dolucan, J.; Mead, D.; Oliver, K.; Omer, A.D.; et al. The gene-rich genome of the scallop Pecten maximus. GigaScience 2020, 9, 9. [Google Scholar] [CrossRef]
- Gerdol, M.; Gomez-Chiarri, M.; Castillo, M.G.; Figueras, A.; Fiorito, G.; Moreira, R.; Novoa, B.; Pallavicini, A.; Ponte, G.; Roumbedakis, K.; et al. Immunity in Molluscs: Recognition and Effector Mechanisms, with a Focus on Bivalvia. In Advances in Comparative Immunology; Springer International Publishing: Cham, Switzerland, 2018; pp. 225–341. ISBN 978-3-319-76768-0. [Google Scholar]
- Hégaret, H.; da Silva, P.M.; Wikfors, G.H.; Haberkorn, H.; Shumway, S.E.; Soudant, P. In vitro interactions between several species of harmful algae and haemocytes of bivalve molluscs. Cell Biol. Toxicol. 2011, 27, 249–266. [Google Scholar] [CrossRef]
- Gerdol, M.; Venier, P. An updated molecular basis for mussel immunity. Fish. Shellfish. Immunol. 2015, 46, 17–38. [Google Scholar] [CrossRef] [Green Version]
- Zhang, G.; Fang, X.; Guo, X.; Li, L.; Luo, R.; Xu, F.; Yang, P.; Zhang, L.; Wang, X.; Qi, H.; et al. The oyster genome reveals stress adaptation and complexity of shell formation. Nat. Cell Biol. 2012, 490, 49–54. [Google Scholar] [CrossRef] [Green Version]
- Cheng, J.; Xun, X.; Kong, Y.; Wang, S.; Yang, Z.; Li, Y.; Kong, D.; Wang, S.; Zhang, L.; Hu, X.; et al. Hsp70 gene expansions in the scallop Patinopecten yessoensis and their expression regulation after exposure to the toxic dinoflagellate Alexandrium catenella. Fish. Shellfish. Immunol. 2016, 58, 266–273. [Google Scholar] [CrossRef]
- Chi, C.; Giri, S.S.; Jun, J.W.; Kim, S.W.; Kim, H.J.; Kang, J.W.; Park, S.C. Detoxification- and Immune-Related Transcriptomic Analysis of Gills from Bay Scallops (Argopecten irradians) in Response to Algal Toxin Okadaic Acid. Toxins 2018, 10, 308. [Google Scholar] [CrossRef] [Green Version]
- Núñez-Acuña, G.; Aballay, A.E.; Hégaret, H.; Astuya, A.P.; Gallardo-Escárate, C. Transcriptional responses of Mytilus chilensis exposed in vivo to saxitoxin (STX). J. Molluscan Stud. 2013, 79, 323–331. [Google Scholar] [CrossRef] [Green Version]
- Cao, R.; Wang, D.; Wei, Q.; Wang, Q.; Yang, D.; Liu, H.; Dong, Z.; Zhang, X.; Zhang, Q.; Zhao, J. Integrative Biomarker Assessment of the Influence of Saxitoxin on Marine Bivalves: A Comparative Study of the Two Bivalve Species Oysters, Crassostrea gigas, and Scallops, Chlamys farreri. Front. Physiol. 2018, 9, 1173. [Google Scholar] [CrossRef] [Green Version]
- Mello, D.F.; de Oliveira, E.S.; Vieira, R.C.; Simões, E.; Trevisan, R.; Dafre, A.L.; Barracco, M.A. Cellular and Transcriptional Responses of Crassostrea gigas Hemocytes Exposed in Vitro to Brevetoxin (PbTx-2). Mar. Drugs 2012, 10, 583–597. [Google Scholar] [CrossRef]
- Pennec, G.L.; Pennec, M.L.; Beninger, G. Seasonal Digestive Gland Dynamics of the Scallop Pecten Maximus in the Bay of Brest (France). J. Mar. Biol. Assoc. UK 2001, 81, 663–671. [Google Scholar] [CrossRef]
- Henry, M.; Boucaud-Camou, E.; Lefort, Y. Functional micro-anatomy of the digestive gland of the scallop Pecten maximus (L.). Aquat. Living Resour. 1991, 4, 191–202. [Google Scholar] [CrossRef]
- Ricard-Blum, S. The Collagen Family. Cold Spring Harb. Perspect. Biol. 2011, 3, a004978. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stobo, L.A.; Lacaze, J.-P.; Scott, A.C.; Petrie, J.; Turrell, E.A. Surveillance of algal toxins in shellfish from Scottish waters. Toxicon 2008, 51, 635–648. [Google Scholar] [CrossRef] [PubMed]
- Schulz, M.H.; Zerbino, D.R.; Vingron, M.; Birney, E. Oases: Robust de novo RNA-seq assembly across the dynamic range of expression levels. Bioinformatics 2012, 28, 1086–1092. [Google Scholar] [CrossRef] [PubMed]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seppey, M.; Manni, M.; Zdobnov, E.M. BUSCO: Assessing Genome Assembly and Annotation Completeness. In Gene Prediction: Methods and Protocols; Methods in Molecular Biology; Kollmar, M., Ed.; Springer: New York, NY, USA, 2019; pp. 227–245. ISBN 978-1-4939-9173-0. [Google Scholar]
- Conesa, A.; Götz, S.; García-Gómez, J.M.; Terol, J.; Talón, M.; Robles, M. Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 2005, 21, 3674–3676. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Götz, S.; García-Gómez, J.M.; Terol, J.; Williams, T.D.; Nagaraj, S.H.; Nueda, M.J.; Robles, M.; Talón, M.; Dopazo, J.; Conesa, A. High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res. 2008, 36, 3420–3435. [Google Scholar] [CrossRef] [PubMed]
- Kriventseva, E.V.; Kuznetsov, D.; Tegenfeldt, F.; Manni, M.; Dias, R.; A Simão, F.; Zdobnov, E.M. OrthoDB v10: Sampling the diversity of animal, plant, fungal, protist, bacterial and viral genomes for evolutionary and functional annotations of orthologs. Nucleic Acids Res. 2019, 47, D807–D811. [Google Scholar] [CrossRef] [Green Version]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [Green Version]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Wang, S.; Zhang, J.; Jiao, W.; Li, J.; Xun, X.; Sun, Y.; Guo, X.; Huan, L.; Dong, B.; Zhang, L.; et al. Scallop genome provides insights into evolution of bilaterian karyotype and development. Nat. Ecol. Evol. 2017, 1, 120. [Google Scholar] [CrossRef]
- Moriya, Y.; Itoh, M.; Okuda, S.; Yoshizawa, A.C.; Kanehisa, M. KAAS: An automatic genome annotation and pathway reconstruction server. Nucleic Acids Res. 2007, 35, W182–W185. [Google Scholar] [CrossRef] [Green Version]
- Szklarczyk, D.; Franceschini, A.; Wyder, S.; Forslund, K.; Heller, D.; Huerta-Cepas, J.; Simonovic, M.; Roth, A.; Santos, A.; Tsafou, K.P.; et al. STRING v10: Protein–protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015, 43, D447–D452. [Google Scholar] [CrossRef]
- Altschul, S.F.; Madden, T.L.; Schäffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef] [Green Version]
- Fisher, R.A. On the Interpretation of χ 2 from Contingency Tables, and the Calculation of P. J. R. Stat. Soc. 1922, 85, 87. [Google Scholar] [CrossRef]
- Andersen, C.L.; Jensen, J.L.; Ørntoft, T.F. Normalization of Real-Time Quantitative Reverse Transcription-PCR Data: A Model-Based Variance Estimation Approach to Identify Genes Suited for Normalization, Applied to Bladder and Colon Cancer Data Sets. Cancer Res. 2004, 64, 5245–5250. [Google Scholar] [CrossRef] [Green Version]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The MIQE Guidelines: Minimum Information for Publication of Quantitative Real-Time PCR Experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef] [Green Version]
- Pfaffl, M.W.; Tichopad, A.; Prgomet, C.; Neuvians, T.P. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper—Excel-based tool using pair-wise correlations. Biotechnol. Lett. 2004, 26, 509–515. [Google Scholar] [CrossRef]
- Vandesompele, J.; de Preter, K.; Pattyn, F.; Poppe, B.; van Roy, N.; de Paepe, A.; Speleman, F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002, 3. [Google Scholar] [CrossRef] [Green Version]
- Volland, M.; Blasco, J.; Hampel, M. Validation of reference genes for RT-qPCR in marine bivalve ecotoxicology: Systematic review and case study using copper treated primary Ruditapes philippinarum hemocytes. Aquat. Toxicol. 2017, 185, 86–94. [Google Scholar] [CrossRef]
- Zhao, S.; Fernald, R.D. Comprehensive Algorithm for Quantitative Real-Time Polymerase Chain Reaction. J. Comput. Biol. J. Comput. Mol. Cell Biol. 2005, 12, 1047–1064. [Google Scholar] [CrossRef] [PubMed]




| Domoic Acid Concentration (µg g−1 Wet Weight): Mean ± SD | ||||||
| DG | Kidney | Gonad | AM | Gill | Other | |
| Control | 2222.9 ± 551.7 | 370.7 ± 238.2 | 39.0 ± 27.2 | 35.1 ± 44.9 | 3.9 ± 3.1 | 14.4 ± 7.0 |
| Treated | 1987.0 ± 406.5 | 1226.3 ± 1037.3 | 42.9 ± 17.7 | 22.9 ± 17.0 | 6.1 ± 6.9 | 14.7 ± 8.8 |
| Domoic Acid Burden (µg): Mean ± SD | ||||||
| DG | Kidney | Gonad | AM | Gill | Other | |
| Control | 3690.2 ± 1002.5 | 88.8 ± 48.2 | 51.2 ± 31.4 | 385.6 ± 526.9 | 8.7 ± 6.5 | 114.2 ± 56.6 |
| Treated | 3884.8 ± 988.6 | 329.0 ± 293.3 | 64.9 ± 30.8 | 262.4 ± 191.2 | 13.2 ± 12.7 | 123.8 ± 62.6 |
| Contig N50 Length | 1883 bp |
| Minimum contig length | 450 bp |
| Maximum contig length | 18,874 bp |
| Average contig length | 1481 bp |
| Total length in contigs | 107,645,527 bp |
| Number of assembled unigenes | 72,673 |
| Sequence ID | Description | FC | padj |
|---|---|---|---|
| ci|000200864|proj|Sample_C_D|2 | cytochrome P450 2D10-like | 16.57 | 5.88 × 10−6 |
| ci|000249823|proj|Sample_C_D|2 | probable proline iminopeptidase | 14.85 | 1.33 × 10−5 |
| ci|000199464|proj|Sample_C_D|2 | pyrroline-5-carboxylate reductase 2-like | 2.68 | 1.57 × 10−5 |
| ci|000262059|proj|Sample_C_D|2 | integumentary mucin C.1-like | 16.32 | 2.09 × 10−5 |
| ci|000180736|proj|Sample_C_D|2 | exocyst complex component 4-like | 2.67 | 2.82 × 10−5 |
| ci|000220117|proj|Sample_C_D|2 | multiple epidermal growth factor-like domains protein 10 | 14.30 | 7.17 × 10−5 |
| ci|000029875|proj|Sample_C_D|2 | cytochrome P450 4F6-like isoform X1 | 2.48 | 1.23 × 10−4 |
| ci|000033337|proj|Sample_C_D|2 | uncharacterized protein LOC117330985 | 4.40 | 1.90 × 10−4 |
| ci|000031940|proj|Sample_C_D|2 | retinol dehydrogenase 7-like isoform X2 | 2.51 | 2.59 × 10−4 |
| ci|000050118|proj|Sample_C_D|2 | 4-coumarate--CoA ligase 1-like | 2.34 | 3.48 × 10−4 |
| ci|000208120|proj|Sample_C_D|2 | 46 kDa FK506-binding nuclear protein-like | 10.84 | 3.73 × 10−4 |
| ci|000199228|proj|Sample_C_D|2 | cholecystokinin receptor-like | 11.25 | 4.78 × 10−4 |
| ci|000033677|proj|Sample_C_D|2 | ectonucleotide pyrophosphatase/phosphodiesterase family member 5-like isoform X2 | 8.80 | 7.96 × 10−4 |
| ci|000181387|proj|Sample_C_D|2 | sodium- and chloride-dependent glycine transporter 1-like | 11.00 | 8.03 × 10−4 |
| ci|000193220|proj|Sample_C_D|2 | uncharacterized protein LOC117327200 | 10.18 | 1.27 × 10−3 |
| ci|000201571|proj|Sample_C_D|2 | uncharacterized protein LOC117327200 | 10.47 | 1.27 × 10−3 |
| ci|000216669|proj|Sample_C_D|2 | NPC intracellular cholesterol transporter 2-like | 9.15 | 1.48 × 10−3 |
| ci|000211651|proj|Sample_C_D|2 | CD109 antigen-like isoform X1 | 3.45 | 1.89 × 10−3 |
| ci|000199247|proj|Sample_C_D|2 | uncharacterized protein LOC117331788 | 5.65 | 1.93 × 10−3 |
| ci|000027261|proj|Sample_C_D|2 | zygotic DNA replication licensing factor mcm3-like | 2.31 | 2.16 × 10−3 |
| Sequence ID | Description | FC | padj |
|---|---|---|---|
| ci|000057484|proj|Sample_C_D|2 | uncharacterized protein LOC110456411 isoform X5 | −56.49 | 1.06 × 10−16 |
| ci|000056978|proj|Sample_C_D|2 | uncharacterized protein LOC110456411 isoform X4 | −53.9 | 7.97 × 10−16 |
| ci|000094075|proj|Sample_C_D|2 | pinin-like | −29.78 | 2.37 × 10−11 |
| ci|000008473|proj|Sample_C_D|2 | uncharacterized protein LOC117340902 | −3.381 | 3.65 × 10−7 |
| ci|000008352|proj|Sample_C_D|2 | uncharacterized protein LOC117325809 isoform X1 | −4.911 | 3.83 × 10−7 |
| ci|000077317|proj|Sample_C_D|2 | uncharacterized protein LOC117338873 | −11.63 | 4.35 × 10−7 |
| ci|000097219|proj|Sample_C_D|2 | serine protease inhibitor Cvsi-1-like | −16.21 | 3.09 × 10−6 |
| ci|000161872|proj|Sample_C_D|2 | uncharacterized protein LOC117338873 | −14.53 | 2.04 × 10−5 |
| ci|000090287|proj|Sample_C_D|2 | innexin unc-7-like | −5.491 | 2.04 × 10−5 |
| ci|000055679|proj|Sample_C_D|2 | putative nuclease HARBI1 | −15.75 | 2.44 × 10−5 |
| ci|000057152|proj|Sample_C_D|2 | uncharacterized protein LOC117338914 | −11.88 | 3.84 × 10−5 |
| ci|000193936|proj|Sample_C_D|2 | uncharacterized protein LOC117332862, partial | −7.373 | 7.17 × 10−5 |
| ci|000071556|proj|Sample_C_D|2 | serine/threonine-protein kinase PINK1, mitochondrial-like | −13.21 | 1.65 × 10−4 |
| ci|000038909|proj|Sample_C_D|2 | sphingomyelin synthase-related protein 1-like | −4.889 | 2.27 × 10−4 |
| ci|000061087|proj|Sample_C_D|2 | uncharacterized protein LOC110461911 isoform X1 | −10.47 | 2.35 × 10−4 |
| ci|000058655|proj|Sample_C_D|2 | apoptosis-stimulating of p53 protein 1-like | −3.122 | 3.73 × 10−4 |
| ci|000104877|proj|Sample_C_D|2 | uncharacterized protein LOC117339040 | −9.715 | 3.90 × 10−4 |
| ci|000017126|proj|Sample_C_D|2 | uncharacterized protein LOC117326392 | −3.294 | 4.56 × 10−4 |
| ci|000019333|proj|Sample_C_D|2 | selenoprotein N-like isoform X1 | −1.813 | 5.94 × 10−4 |
| ci|000009958|proj|Sample_C_D|2 | uncharacterized protein LOC117345233 | −2.103 | 6.09 × 10−4 |
| Sequence ID | Description | Type | FC | padj |
|---|---|---|---|---|
| ci|000016850|proj|Sample_C_D|2 | XP_033740949.1 metabotropic glutamate receptor 1-like [Pecten maximus] | Metabotropic | 2.938 | 0.278 |
| ci|000042484|proj|Sample_C_D|2 | XP_033740949.1 metabotropic glutamate receptor 1-like [Pecten maximus] | Metabotropic | 1.309 | 0.937 |
| ci|000199286|proj|Sample_C_D|2 | XP_033740949.1 metabotropic glutamate receptor 1-like [Pecten maximus] | Metabotropic | 2.566 | 0.288 |
| ci|000235714|proj|Sample_C_D|2 | XP_033740949.1 metabotropic glutamate receptor 1-like [Pecten maximus] | Metabotropic | 1.575 | NA |
| ci|000005744|proj|Sample_C_D|2 | XP_033744387.1 glutamate receptor-like [Pecten maximus] | Ionotropic | 1.504 | 0.714 |
| ci|000012480|proj|Sample_C_D|2 | XP_033744387.1 glutamate receptor-like [Pecten maximus] | Ionotropic | 1.567 | 0.765 |
| ci|000178783|proj|Sample_C_D|2 | XP_033744387.1 glutamate receptor-like [Pecten maximus] | Ionotropic | 3.428 | 0.123 |
| ci|000194581|proj|Sample_C_D|2 | XP_033744387.1 glutamate receptor-like [Pecten maximus] | Ionotropic | 1.655 | 0.845 |
| ci|000000691|proj|Sample_C_D|2 | XP_033760483.1 glutamate receptor 3-like [Pecten maximus] | Ionotropic | 1.242 | 0.945 |
| ci|000077591|proj|Sample_C_D|2 | XP_033760483.1 glutamate receptor 3-like [Pecten maximus] | Ionotropic | −1.006 | 0.998 |
| ci|000108874|proj|Sample_C_D|2 | XP_033760483.1 glutamate receptor 3-like [Pecten maximus] | Ionotropic | −1.199 | 0.968 |
| ci|000219160|proj|Sample_C_D|2 | XP_033760483.1 glutamate receptor 3-like [Pecten maximus] | Ionotropic | −1.319 | 0.937 |
| ci|000054856|proj|Sample_C_D|2 | XP_033760586.1 glutamate receptor U1-like [Pecten maximus] | Ionotropic | −1.100 | 0.970 |
| ci|000106439|proj|Sample_C_D|2 | XP_033760586.1 glutamate receptor U1-like [Pecten maximus] | Ionotropic | −1.447 | 0.912 |
| ci|000013492|proj|Sample_C_D|2 | XP_033763623.1 metabotropic glutamate receptor 7-like isoform X3 [Pecten maximus] | Metabotropic | 1.042 | 0.985 |
| ci|000015386|proj|Sample_C_D|2 | XP_033763623.1 metabotropic glutamate receptor 7-like isoform X3 [Pecten maximus] | Metabotropic | 1.066 | NA |
| ci|000028888|proj|Sample_C_D|2 | XP_033763623.1 metabotropic glutamate receptor 7-like isoform X3 [Pecten maximus] | Metabotropic | 1.169 | 0.937 |
| ci|000048793|proj|Sample_C_D|2 | XP_033763623.1 metabotropic glutamate receptor 7-like isoform X3 [Pecten maximus] | Metabotropic | 1.161 | 0.928 |
| ci|000191381|proj|Sample_C_D|2 | XP_033763623.1 metabotropic glutamate receptor 7-like isoform X3 [Pecten maximus] | Metabotropic | 1.348 | 0.937 |
| Functional Annotation | Number | % |
|---|---|---|
| Differentially expressed unigenes | 535 | 100 |
| With Blastx hit | 286 | 53.5 |
| With GO terms | 259 | 48.4 |
| With enzyme code | 129 | 24.1 |
| With KO ortholog | 102 | 19.1 |
| With PFAM domains | 188 | 35.1 |
| All unigenes | 72,673 | 100 |
| With Blastx hit | 35,583 | 49.0 |
| With GO terms | 32,203 | 44.3 |
| With enzyme code | 16,429 | 22.6 |
| With KO ortholog | 7657 | 10.5 |
| With PFAM domains | 24,810 | 34.1 |
| Sequence Name | Description | Symbol | Sense Primer | Antisense Primer | bp | E |
|---|---|---|---|---|---|---|
| ci|000200326|proj|Sample_C_D|2 | glyceraldehyde-3-phosphate dehydrogenase | GAPDH | TCCGGATGTGTCTGTTGTTGAC | TTCAGATCTCCATCAGCTGCAC | 102 | 0.8465 |
| ci|000017232|proj|Sample_C_D|2 | eukaryotic translation elongation factor 1 alpha | EF1A | AGGGCTCCTTCAAGTATGCCTG | TGAGCGGTCTCGAACTTCCAC | 100 | 0.8275 |
| ci|000019926|proj|Sample_C_D|2 | cytochrome c oxidase subunit 1 | COX1 | AGTGGAGAACTATTGGGTGTGC | AGACCTAGGCCGATTTCCAAAC | 119 | 0.8525 |
| ci|000005190|proj|Sample_C_D|2 | NADH dehydrogenase (ubiquinone) 1 alpha subcomplex subunit 7 | NDUFA7 | ATTACACACGAGATGGACGCTG | ACATCAGAGCTGGCTGTTTCAG | 115 | 0.8043 |
| ci|000002411|proj|Sample_C_D|2 | multidrug resistance-associated protein 7 | MRP7 | CGGATGGTGGCTAACTCATT | CGATGCACCCATACACTGTC | 200 | 0.8594 |
| ci|000062123|proj|Sample_C_D|2 | cytochrome p450 2b4-like | CYP2B4 | CATGCGAAGGACTACGACAAG | GAACAAAATGGCCCAAAGAAG | 183 | 0.8314 |
| ci|000199464|proj|Sample_C_D|2 | pyrroline-5-carboxylate reductase 2-like | P5CR | CCTCACATCATCACTCCA GTCC | GACGGGAGCAGATTCTCCTC | 119 | 0.8547 |
| ci|000181387|proj|Sample_C_D|2 | sodium- and chloride-dependent glycine transporter 1-like | SLC6A9 | GACGGTACTGGGCATTTCTG | ATCAGCAAGGCCGTAAGGAG | 183 | 0.8539 |
| ci|000018690|proj|Sample_C_D|2 | ferritin 2 | FERRITIN | CCATGCTGAAACCGAGGCTG | CAATCCTGCCTCCTCTCTTG | 206 | 0.8547 |
| ci|000200864|proj|Sample_C_D|2 | cytochrome p450 2u1-like | CYP2U1 | CATCGACGCCTTCCAGTTCG | GATAGTCAGCCATCGTGGGT | 233 | 0.8753 |
| Rank | geNorm (Average M) | NormFinder (Stability) | BestKeeper (r) | BestKeeper (SD) | ||||
|---|---|---|---|---|---|---|---|---|
| 1 | GAPDH-COX1 | 0.78 | GAPDH | 0.105 | COX1 | 0.858 | GAPDH | 0.724 |
| 2 | GAPDH-COX1 | 0.78 | COX1 | 0.196 | GAPDH | 0.782 | NDUFA7 | 0.781 |
| 3 | EF1A | 1.01 | EF1A | 0.244 | EF1A | 0.662 | COX1 | 0.838 |
| 4 | NDUFA7 | 1.34 | NDUFA7 | 0.386 | NDUFA7 | −0.243 | EF1A | 0.861 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ventoso, P.; Pazos, A.J.; Blanco, J.; Pérez-Parallé, M.L.; Triviño, J.C.; Sánchez, J.L. Transcriptional Response in the Digestive Gland of the King Scallop (Pecten maximus) After the Injection of Domoic Acid. Toxins 2021, 13, 339. https://doi.org/10.3390/toxins13050339
Ventoso P, Pazos AJ, Blanco J, Pérez-Parallé ML, Triviño JC, Sánchez JL. Transcriptional Response in the Digestive Gland of the King Scallop (Pecten maximus) After the Injection of Domoic Acid. Toxins. 2021; 13(5):339. https://doi.org/10.3390/toxins13050339
Chicago/Turabian StyleVentoso, Pablo, Antonio J. Pazos, Juan Blanco, M. Luz Pérez-Parallé, Juan C. Triviño, and José L. Sánchez. 2021. "Transcriptional Response in the Digestive Gland of the King Scallop (Pecten maximus) After the Injection of Domoic Acid" Toxins 13, no. 5: 339. https://doi.org/10.3390/toxins13050339
APA StyleVentoso, P., Pazos, A. J., Blanco, J., Pérez-Parallé, M. L., Triviño, J. C., & Sánchez, J. L. (2021). Transcriptional Response in the Digestive Gland of the King Scallop (Pecten maximus) After the Injection of Domoic Acid. Toxins, 13(5), 339. https://doi.org/10.3390/toxins13050339