Honeybee Venom Proteome Profile of Queens and Winter Bees as Determined by a Mass Spectrometric Approach
Abstract
:1. Introduction
2. Materials and Methods
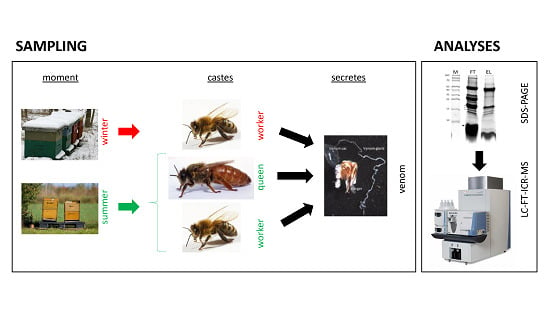
2.1. Venom Collection
2.2. Mass Spectrometric Analysis
2.3. Criteria for Positive Identifications
2.4. Sequence Analysis
3. Results and Discussion
3.1. Identification and Categorization of Venom Proteins

3.2. Comparison of the Venom Composition between Summer and Winter Worker Honeybees
| Name | Acc. N° | Allergen | SignalP | EX | Summer Worker | Winter Worker | Queen | Similar Venom Compound | ||
|---|---|---|---|---|---|---|---|---|---|---|
| Species | Evidence | Ref. | ||||||||
| A. Putative toxins | ||||||||||
| Esterases | ||||||||||
| Phospholipase A2-1 | gi|58585172 | Api m 1 | x | - | x | x | x | - | - | - |
| Phospholipase A2-2 | gi|110758297 | - | x | - | x | x | x | - | - | - |
| Group XV phospholipase A2 | gi|328791555 | - | x | - | x | - | - | Ophiophagus hannah | P+T | [20] |
| Acid phosphatase | gi|301601654 | Api m 3 | x | x | x | x | x | Nasonia vitripennis | P | [21] |
| Acid phosphatase 2 | gi|571531089 | - | x | x | x | x | x | Pteromalus puparum | T+EA | [22] |
| 5′-Nucleotidase | gi|66523706 | - | x | x | x | x | Gloydius blomhoffi | P | [23] | |
| Carboxylesterase | gi|187281550 | Api m 8 | x | x | x | x | Nasonia vitripennis | P | [21] | |
| Proteases and peptidases | ||||||||||
| CLIP serine protease | gi|571522677 | - | x | - | x | x | x | Bombus ignitus | P | [24] |
| CUB serine protease 1 | gi|58585116 | Api m 7 | x | - | x | x | x | Nasonia vitripennis | T | [21] |
| CUB serine protease 2 | gi|48101366 | - | x | - | x | x | x | Nasonia vitripennis | T | [21] |
| Putative trypsin | au9.g8903.t1 | - | x | - | x | - | x | Nasonia vitripennis | P | [21] |
| Serine protease snake | gi|328783264 | - | x | - | x | x | - | Ophiophagus hannah | P+T | [20] |
| Dipeptidyl peptidase IV | gi|187281543 | Api m 5 | x | x | x | x | x | Vespula vulgaris | P | [25] |
| Serine carboxypeptidase | gi|226533687 | Api m 9 | x | x | x | x | x | Croatlus adamanteus | T | [26] |
| Prolylcarboxypeptidase | gi|328778095 | - | x | x | x | x | x | Ophiophagus hannah | P+T | [20] |
| Metalloprotease | gi|571501445 | - | x | x | x | x | x | Eulophus pennicornis | [27] | |
| Serine proteinase stubble | au9.g5504.t1 | - | x | - | - | - | x | Crotalus adamanteus | T | [26] |
| Protease inhibitors | ||||||||||
| Api m 6 | gi|94400907 | Api m 6 | x | x | x | x | - | - | - | |
| Serpin 1 | gi|328793022 | - | x | x | x | x | x | Ophiophagus hannah | P+T | [20] |
| Serpin 2 | gi|328791596 | - | x | x | x | x | x | Tityus bahiensis | T | [28] |
| Serpin 3 | gi|328780925 | - | x | x | x | x | Tityus bahiensis | T | [28] | |
| Antithrombin-III | gi|571552510 | - | x | x | - | - | x | Tityus bahiensis | T | [28] |
| Carbohydrate metabolism | ||||||||||
| Hyaluronidase | gi|58585182 | Api m 2 | x | - | x | x | x | Apis cerana | T | [29] |
| N-sulfoglucosamine sulfohydrolase | gi|328793712 | - | x | - | x | x | x | Boiga irregularis | T | [30] |
| Endochitinase | gi|66511507 | - | x | - | x | x | x | Nasonia vitripennis | T | [21] |
| Growth factors | ||||||||||
| Platelet-derived growth factor | gi|571515288 | - | x | x | x | x | x | Echis coloratus | T | [31] |
| Imaginal disc growth factor 4 | gi|571545715 | - | x | - | x | x | x | Chelonus inanitus | P | [32] |
| Major royal jelly proteins | ||||||||||
| MRJP8 | gi|58585070 | Api m 11.0101 | x | - | x | x | x | Chelonus inanitus | P | [32] |
| MRJP9 | gi|67010041 | Api m 11.0201 | x | - | x | x | x | Chelonus inanitus | P | [32] |
| Peptides | ||||||||||
| Melittin | gi|58585154 | Api m 4 | x | - | x | x | x | Vespula maculifrons | T | [33] |
| Apamin | gi|58585166 | - | x | - | x | x | x | - | - | - |
| Secapin | gi|58585180 | - | x | - | x | x | x | Vespa velutina nigrithorax | U | gi|33321084 |
| Other toxins | ||||||||||
| C-type lectin | gi|328792562 | - | x | - | x | - | - | - | - | - |
| Icarapin | gi|60115688 | Api m 10 | x | - | x | x | x | Apis cerana | T | [34] |
| B.Selected trace molecules | ||||||||||
| Secreted proteins | ||||||||||
| C1q-like protein | gi|221325614 | - | x | x | x | x | x | Nasonia vitripennis | P | [21] |
| Lysozyme c-1 | gi|506614822 | - | x | x | x | x | - | Ophiophagus hannah | P+T | [20] |
| Peptidoglycan-recognition protein SA | gi|254910928 | - | x | x | x | x | - | Ophiophagus hannah | P+T | [20] |
| Transferrin | gi|58585086 | - | x | x | x | - | - | Crotalus horridus | T | [35] |
| Modular serine protease | gi|328780689 | - | x | - | x | x | - | Ophiophagus hannah | P+T | [20] |
| Cathepsin F | gi|328788558 | - | x | - | x | x | x | Tityus bahiensis | T | [28] |
| Cathepsin K | au9.g225.t1 | - | x | - | x | x | - | Micrurus fulvius | T | [36] |
| Peptidylglycine α-hydroxylating monooxygenase | gi|328787622 | - | x | x | x | x | x | Boiga irregularis | T | [30] |
| Apolipophorins | gi|571543905 | - | x | x | x | x | - | Ophiophagus hannah | P+T | [20] |
| Dorsal-ventral patterning protein Sog | gi|328791019 | - | x | - | x | - | - | Ophiophagus hannah | P+T | [20] |
| Laminin subunit γ-1 | gi|571556732 | - | x | x | x | x | x | Ophiophagus hannah | P+T | [20] |
| Vitellogenin | gi|58585104 | Api m 12 | x | (x) | - | x | Bombus terrestris | P | [37] | |
| Glutathione S-transferase | gi|571577571 | - | x | - | - | x | Boiga irregularis | T | [30] | |
| Protein kinase C-binding protein NELL1 | au9.g225.t1 | - | x | - | - | - | x | Ophiophagus hannah | P+T | [20] |
| Odorant binding protein 14 | au9.g8525.t1 | - | x | - | (x) | x | - | - | - | - |
| Aldose 1-epimerase | gi|66541614 | - | x | x | - | x | - | Boiga irregularis | T | [30] |
| poly(U)-specific endoribonuclease | gi|110760204 | - | x | - | - | x | - | Ophiophagus hannah | P+T | [20] |
| Peritrophins 3-B | gi|288869483 | - | x | - | - | x | - | - | - | - |
3.3. Comparison of the Venom Composition between Honeybee Workers and Queens
3.4. Comparison of Honeybee Venom Allergens
4. Conclusions
Supplementary Files
Supplementary File 1Acknowledgments
Author Contributions
Conflicts of Interest
References
- Fry, B.G.; Roelants, K.; Champagne, D.E.; Scheib, H.; Tyndall, J.D.; King, G.F.; Nevalainen, T.J.; Norman, J.A.; Lewis, R.J.; Norton, R.S.; et al. The toxicogenomic multiverse: Convergent recruitment of proteins into animal venoms. Annu. Rev. Genomics Hum. Genet. 2009, 10, 483–511. [Google Scholar] [CrossRef] [PubMed]
- Leluk, J.; Schmidt, J.; Jones, D. Comparative studies on the protein composition of hymenopteran venom reservoirs. Toxicon 1989, 27, 105–114. [Google Scholar] [CrossRef]
- Resende, V.M.; Vasilj, A.; Santos, K.S.; Palma, M.S.; Shevchenko, A. Proteome and phosphoproteome of Africanized and European honeybee venoms. Proteomics. 2013, 13, 2638–2648. [Google Scholar] [CrossRef] [PubMed]
- Wilson, E.O.; Holldobler, B. Eusociality: Origin and consequences. Proc. Natl. Acad. Sci. USA 2005, 102, 13367–13371. [Google Scholar] [CrossRef] [PubMed]
- Eliyahu, D.; Ross, K.G.; Haight, K.L.; Keller, L.; Liebig, J. Venom alkaloid and cuticular hydrocarbon profiles are associated with social organization, queen fertility status, and queen genotype in the fire ant Solenopsis invicta. J. Chem. Ecol. 2011, 37, 1242–1254. [Google Scholar] [CrossRef] [PubMed]
- Tschinkel, W.R. The Fire Ants; Harvard University Press: Cambridge, England, 2006. [Google Scholar]
- Schmidt, J.O. Toxinology of venoms from the honeybee genus. Apis. Toxicon 1995, 33, 917–927. [Google Scholar] [CrossRef]
- Owen, M.D. Relationship between age and hyaluronidase activity in the venom of queen and worker honey bees (Apis mellifera L.). Toxicon 1979, 17, 94–98. [Google Scholar] [CrossRef]
- Vlasak, R.; Kreil, G. Nucleotide sequence of cloned cDNAs coding for preprosecapin, a major product of queen-bee venom glands. Eur. J. Biochem. 1984, 145, 279–282. [Google Scholar] [CrossRef] [PubMed]
- Nocelli, R.C.F.; Roat, T.C.; Cruz-Landim, C. Alterations induced by the juvenile hormone in glandular cells of the Apis mellifera venom gland: Applications on newly emerged workers (Hymenoptera, Apidae). Micron 2007, 38, 74–80. [Google Scholar] [CrossRef] [PubMed]
- De Abreu, R.M.; Silva de Moraes, R.L.; Camargo-Mathias, M.I. Biochemical and cytochemical studies of the enzymatic activity of the venom glands of workers of honey bee Apis mellifera L. (Hymenoptera, Apidae). Micron 2010, 41, 172–175. [Google Scholar] [CrossRef] [PubMed]
- Cologna, C.T.; Cardoso, J.S.; Jourdan, E.; Degueldre, M.; Upert, G.; Gilles, N.; Uetanabaro, A.P.; Costa Neto, E.M.; Thonart, P.; de, P.E.; et al. Peptidomic comparison and characterization of the major components of the venom of the giant ant Dinoponera quadriceps collected in four different areas of Brazil. J. Proteomics 2013, 94, 413–422. [Google Scholar] [CrossRef] [PubMed]
- Brand, J.M.; Blum, M.S.; Barlin, M.R. Fire ant venoms: Intraspecific and interspecific variation among castes and individuals. Toxicon 1973, 11, 325–331. [Google Scholar] [CrossRef]
- Lai, L.C.; Hua, K.H.; Yang, C.C.; Huang, R.N.; Wu, W.J. Secretion profiles of venom alkaloids in Solenopsis geminata (Hymenoptera: Formicidae) in Taiwan. Environ. Entomol 2009, 38, 879–884. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, R.S., Jr.; Sciani, J.M.; Marques-Porto, R.; Junior, A.L.; Orsi, R.O.; Barraviera, B.; Pimenta, D.C. Africanized honey bee (Apis mellifera) venom profiling: Seasonal variation of melittin and phospholipase A2 levels. Toxicon 2010, 56, 355–362. [Google Scholar] [CrossRef] [PubMed]
- Van Vaerenbergh, M.; Cardoen, D.; Formesyn, E.M.; Brunain, M.; van, D.G.; Blank, S.; Spillner, E.; Verleyen, P.; Wenseleers, T.; Schoofs, L.; et al. Extending the honey bee venome with the antimicrobial peptide apidaecin and a protein resembling wasp antigen 5. Insect Mol. Biol. 2013, 22, 199–210. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Colinet, D.; Deleury, E.; Anselme, C.; Cazes, D.; Poulain, J.; Azema-Dossat, C.; Belghazi, M.; Gatti, J.L.; Poirie, M. Extensive inter- and intraspecific venom variation in closely related parasites targeting the same host: the case of Leptopilina parasitoids of Drosophila. Insect Biochem. Mol. Biol. 2013, 43, 601–611. [Google Scholar] [CrossRef] [PubMed]
- Van Vaerenbergh, M.; Debyser, G.; Devreese, B.; de Graaf, D.C. Exploring the hidden honeybee (Apis mellifera) venom proteome by integrating a combinatorial peptide ligand library approach with FTMS. J. Proteomics. 2014, 99, 169–178. [Google Scholar] [CrossRef] [PubMed]
- Peiren, N.; Vanrobaeys, F.; de Graaf, D.C.; Devreese, B.; Van, B.J.; Jacobs, F.J. The protein composition of honeybee venom reconsidered by a proteomic approach. Biochim. Biophys. Acta 2005, 1752, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Vonk, F.J.; Casewell, N.R.; Henkel, C.V.; Heimberg, A.M.; Jansen, H.J.; McCleary, R.J.; Kerkkamp, H.M.; Vos, R.A.; Guerreiro, I.; Calvete, J.J.; et al. The king cobra genome reveals dynamic gene evolution and adaptation in the snake venom system. Proc. Natl. Acad. Sci. USA 2013, 110, 20651–20656. [Google Scholar] [CrossRef] [PubMed]
- De Graaf, D.C.; Aerts, M.; Brunain, M.; Desjardins, C.A.; Jacobs, F.J.; Werren, J.H.; Devreese, B. Insights into the venom composition of the ectoparasitoid wasp Nasonia vitripennis from bioinformatic and proteomic studies. Insect Mol. Biol. 2010, 19 (Suppl 1), 11–26. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.Y.; Ye, G.Y.; Hu, C. Molecular cloning and characterization of acid phosphatase in venom of the endoparasitoid wasp Pteromalus puparum (Hymenoptera: Pteromalidae). Toxicon 2008, 51, 1391–1399. [Google Scholar] [CrossRef] [PubMed]
- Ogawa, Y.; Murayama, N.; Yanoshita, R. Molecular cloning and characterization of ecto-5'-nucleotidase from the venoms of Gloydius blomhoffi. Toxicon 2009, 54, 408–412. [Google Scholar] [CrossRef] [PubMed]
- Choo, Y.M.; Lee, K.S.; Yoon, H.J.; Kim, B.Y.; Sohn, M.R.; Roh, J.Y.; Je, Y.H.; Kim, N.J.; Kim, I.; Woo, S.D.; et al. Dual function of a bee venom serine protease: Prophenoloxidase-activating factor in arthropods and fibrin(ogen)olytic enzyme in mammals. PLoS One 2010, 5, e10393. [Google Scholar] [CrossRef] [PubMed]
- Blank, S.; Seismann, H.; Bockisch, B.; Braren, I.; Cifuentes, L.; McIntyre, M.; Ruhl, D.; Ring, J.; Bredehorst, R.; Ollert, M.W.; Grunwald, T.; Spillner, E. Identification, recombinant expression, and characterization of the 100 kDa high molecular weight Hymenoptera venom allergens Api m 5 and Ves v 3. J. Immunol. 2010, 184, 5403–5413. [Google Scholar] [CrossRef] [PubMed]
- Rokyta, D.R.; Lemmon, A.R.; Margres, M.J.; Aronow, K. The venom-gland transcriptome of the eastern diamondback rattlesnake (Crotalus adamanteus). BMC Genomics 2012, 13, 312. [Google Scholar] [CrossRef] [PubMed]
- Price, D.R.; Bell, H.A.; Hinchliffe, G.; Fitches, E.; Weaver, R.; Gatehouse, J.A. A venom metalloproteinase from the parasitic wasp Eulophus pennicornis is toxic towards its host, tomato moth (Lacanobia oleracae). Insect Mol. Biol. 2009, 18, 195–202. [Google Scholar] [CrossRef] [PubMed]
- De Oliveira, U.C.; Candido, D.M.; Dorce, V.A.; Junqueira-de-Azevedo, I.L. The transcriptome recipe for the venom cocktail of Tityus bahiensis scorpion. Toxicon 2015, 95, 52–61. [Google Scholar] [CrossRef] [PubMed]
- Shen, L.R.; Zhan, C.X.; Cheng, J.A. Cloning and sequencing of gene encoding hyaluronidase from the venom of Apis cerana cerana. Entomolia Sin 2001, 8, 353–360. [Google Scholar]
- McGivern, J.J.; Wray, K.P.; Margres, M.J.; Couch, M.E.; Mackessy, S.P.; Rokyta, D.R. RNA-seq and high-definition mass spectrometry reveal the complex and divergent venoms of two rear-fanged colubrid snakes. BMC Genomics 2014, 15, 1061. [Google Scholar] [CrossRef] [PubMed]
- Hargreaves, A.D.; Swain, M.T.; Logan, D.W.; Mulley, J.F. Testing the Toxicofera: Comparative transcriptomics casts doubt on the single, early evolution of the reptile venom system. Toxicon 2014, 92, 140–156. [Google Scholar] [CrossRef] [PubMed]
- Vincent, B.; Kaeslin, M.; Roth, T.; Heller, M.; Poulain, J.; Cousserans, F.; Schaller, J.; Poirie, M.; Lanzrein, B.; Drezen, J.M.; et al. The venom composition of the parasitic wasp Chelonus inanitus resolved by combined expressed sequence tags analysis and proteomic approach. BMC Genomics 2010, 11, 693. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shi, W.J.; Zhang, S.F.; Zhang, C.X.; Cheng, J.A. Cloning and comparative analysis of the venom prepromelittin genes from four wasp species. Yi. Chuan Xue. Bao. 2003, 30, 555–559. [Google Scholar] [PubMed]
- Wong, K.L.; Li, H.; Wong, K.K.; Jiang, T.; Shaw, P.C. Location and reduction of icarapin antigenicity by site specific coupling to polyethylene glycol. Protein Pept. Lett. 2012, 19, 238–243. [Google Scholar] [CrossRef] [PubMed]
- Rokyta, D.R.; Wray, K.P.; Margres, M.J. The genesis of an exceptionally lethal venom in the timber rattlesnake (Crotalus horridus) revealed through comparative venom-gland transcriptomics. BMC Genomics 2013, 14, 394. [Google Scholar] [CrossRef] [PubMed]
- Margres, M.J.; Aronow, K.; Loyacano, J.; Rokyta, D.R. The venom-gland transcriptome of the eastern coral snake (Micrurus fulvius) reveals high venom complexity in the intragenomic evolution of venoms. BMC Genomics 2013, 14, 531. [Google Scholar] [CrossRef] [PubMed]
- Van Vaerenbergh, M.; Debyser, G.; Smagghe, G.; Devreese, B.; de Graaf, D.C. Unraveling the venom proteome of the bumblebee (Bombus terrestris) by integrating a combinatorial peptide ligand library approach with FT-ICR MS. Toxicon 2015, 102, 81–88. [Google Scholar] [CrossRef] [PubMed]
- Kontogiannatos, D.; Michail, X.; Kourti, A. Molecular characterization of an ecdysteroid inducible carboxylesterase with GQSCG motif in the corn borer, Sesamia nonagrioides. J. Insect Physiol. 2011, 57, 1000–1009. [Google Scholar] [CrossRef] [PubMed]
- Blank, S.; Seismann, H.; McIntyre, M.; Ollert, M.; Wolf, S.; Bantleon, F.I.; Spillner, E. Vitellogenins are new high molecular weight components and allergens (Api m 12 and Ves v 6) of Apis mellifera and Vespula vulgaris venom. PLoS One 2013, 8, e62009. [Google Scholar] [CrossRef] [PubMed]
- Engelmann, F. Undegraded vitellogenin polysomes from female insect fat bodies. Biochem. Biophys. Res. Commun. 1977, 78, 641–647. [Google Scholar] [CrossRef]
- Wheeler, D.E.; Kawooya, J.K. Purification and characterization of honey bee vitellogenin. Arch. Insect Biochem. Physiol. 1990, 14, 253–267. [Google Scholar] [CrossRef] [PubMed]
- Barchuk, A.R.; Bitondi, M.M.; Simoes, Z.L. Effects of juvenile hormone and ecdysone on the timing of vitellogenin appearance in hemolymph of queen and worker pupae of Apis mellifera. J. Insect Sci. 2002, 2, 1. [Google Scholar] [CrossRef] [PubMed]
- Engels, W.; aatz, H.; illikens, A.; imoes, Z.L.P.; rube, A.; Braun, R.; Dittrich, F. Honey bee reproduction: Vitellogenin and caste-specificregulation of fertility. In Advances in Invertebrate Reproduction; Hoshi, M., Yamashito, O., Eds.; Elsevier: Amsterdam, The Netherlands, 1990; Volume 5, pp. 495–502. [Google Scholar]
- Fluri, P.; Luscher, M.; Wille, H.; Gerig, L. Changes in weight of the pharyngeal gland and hemolymph titers of juvenile-hormone, protein and vitellogenin in worker honey bees. J. Insect Physiol. 1982, 28, 61–68. [Google Scholar] [CrossRef]
- Amdam, G.V.; Simoes, Z.L.; Hagen, A.; Norberg, K.; Schroder, K.; Mikkelsen, O.; Kirkwood, T.B.; Omholt, S.W. Hormonal control of the yolk precursor vitellogenin regulates immune function and longevity in honeybees. Exp. Gerontol. 2004, 39, 767–773. [Google Scholar] [CrossRef] [PubMed]
- Guidugli, K.R.; Nascimento, A.M.; Amdam, G.V.; Barchuk, A.R.; Omholt, S.; Simoes, Z.L.; Hartfelder, K. Vitellogenin regulates hormonal dynamics in the worker caste of a eusocial insect. FEBS Lett. 2005, 579, 4961–4965. [Google Scholar] [CrossRef] [PubMed]
- Nelson, C.M.; Ihle, K.E.; Fondrk, M.K.; Page, R.E.; Amdam, G.V. The gene vitellogenin has multiple coordinating effects on social organization. PLoS Biol. 2007, 5, e62. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Danneels, E.L.; Van Vaerenbergh, M.; Debyser, G.; Devreese, B.; De Graaf, D.C. Honeybee Venom Proteome Profile of Queens and Winter Bees as Determined by a Mass Spectrometric Approach. Toxins 2015, 7, 4468-4483. https://doi.org/10.3390/toxins7114468
Danneels EL, Van Vaerenbergh M, Debyser G, Devreese B, De Graaf DC. Honeybee Venom Proteome Profile of Queens and Winter Bees as Determined by a Mass Spectrometric Approach. Toxins. 2015; 7(11):4468-4483. https://doi.org/10.3390/toxins7114468
Chicago/Turabian StyleDanneels, Ellen L., Matthias Van Vaerenbergh, Griet Debyser, Bart Devreese, and Dirk C. De Graaf. 2015. "Honeybee Venom Proteome Profile of Queens and Winter Bees as Determined by a Mass Spectrometric Approach" Toxins 7, no. 11: 4468-4483. https://doi.org/10.3390/toxins7114468
APA StyleDanneels, E. L., Van Vaerenbergh, M., Debyser, G., Devreese, B., & De Graaf, D. C. (2015). Honeybee Venom Proteome Profile of Queens and Winter Bees as Determined by a Mass Spectrometric Approach. Toxins, 7(11), 4468-4483. https://doi.org/10.3390/toxins7114468