Neural Circuits on a Chip
Abstract
:1. Introduction
2. Dissociated Neural Cultures
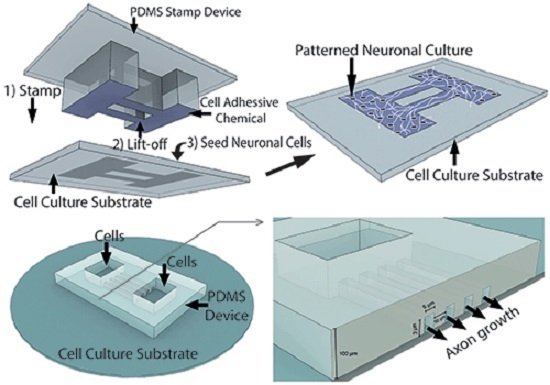
3. Neural Culture Patterning Methods
4. Patterned Networks
5. Neural Circuits Carry Out Logic Functions
6. Structured Organotypic and 3D Dissociated Neural Cultures
7. Interfaces to Neural Circuits
7.1. Electrical Interfaces
7.2. Optical Interfaces
8. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Banker, G.; Goslin, K. Culturing Nerve Cells, 2nd ed.; MIT Press: Cambridge, MA, USA, 1998. [Google Scholar]
- Millet, L.J.; Gillette, M.U. Over a century of neuron culture: from the hanging drop to microfluidic devices. Yale J. Biol. Med. 2012, 85, 501–521. [Google Scholar] [PubMed]
- Nieland, T.J.F.; Logan, D.J.; Saulnier, J.; Lam, D.; Johnson, C.; Root, D.E.; Carpenter, A.E.; Sabatini, B.L. High content image analysis identifies novel regulators of synaptogenesis in a high-throughput RNAi screen of primary neurons. PLoS ONE 2014, 9, e91744. [Google Scholar] [CrossRef] [PubMed]
- Paradis, S.; Harrar, D.B.; Lin, Y.; Koon, A.C.; Hauser, J.L.; Griffith, E.C.; Zhu, L.; Brass, L.F.; Chen, C.; Greenberg, M.E. An RNAi-based approach identifies molecules required for glutamatergic and GABAergic synapse development. Neuron 2007, 53, 217–232. [Google Scholar] [CrossRef] [PubMed]
- Van Pelt, J.; Wolters, P.S.; Corner, M.A.; Rutten, W.L.C.; Ramakers, G.J.A. Long-term characterization of firing dynamics of spontaneous bursts in cultured neural networks. IEEE Trans. Biomed. Eng. 2004, 51, 2051–2062. [Google Scholar] [CrossRef] [PubMed]
- Wagenaar, D.A.; Pine, J.; Potter, S.M. An extremely rich repertoire of bursting patterns during the development of cortical cultures. BMC Neurosci. 2006, 7, 11. [Google Scholar] [CrossRef] [PubMed]
- Eytan, D.; Marom, S. Dynamics and effective topology underlying synchronization in networks of cortical neurons. J. Neurosci. Off. J. Soc. Neurosci. 2006, 26, 8465–8476. [Google Scholar] [CrossRef] [PubMed]
- Ivenshitz, M.; Segal, M. Neuronal density determines network connectivity and spontaneous activity in cultured hippocampus. J. Neurophysiol. 2010, 104, 1052–1060. [Google Scholar] [CrossRef] [PubMed]
- Wyart, C.; Ybert, C.; Bourdieu, L.; Herr, C.; Prinz, C.; Chatenay, D. Constrained synaptic connectivity in functional mammalian neuronal networks grown on patterned surfaces. J. Neurosci. Methods 2002, 117, 123–131. [Google Scholar] [CrossRef]
- Brewer, G.J.; Boehler, M.D.; Leondopulos, S.; Pan, L.; Alagapan, S.; DeMarse, T.B.; Wheeler, B.C. Toward a self-wired active reconstruction of the hippocampal trisynaptic loop: DG-CA3. Front. Neural Circuits 2013, 7, 165. [Google Scholar] [CrossRef] [PubMed]
- Soriano, J.; Rodríguez Martínez, M.; Tlusty, T.; Moses, E. Development of input connections in neural cultures. Proc. Natl. Acad. Sci. USA 2008, 105, 13758–13763. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Huang, Z.; Yang, K.; Liu, W.; Xie, Y.; Yuan, B.; Zhang, W.; Jiang, X. Self-organizing circuit assembly through spatiotemporally coordinated neuronal migration within geometric constraints. PLoS ONE 2011, 6, e28156. [Google Scholar] [CrossRef] [PubMed]
- Brewer, G.J. Isolation and culture of adult rat hippocampal neurons. J. Neurosci. Methods 1997, 71, 143–155. [Google Scholar] [CrossRef]
- Brewer, G.J.; Torricelli, J.R.; Evege, E.K.; Price, P.J. Optimized survival of hippocampal neurons in B27-supplemented Neurobasal, a new serum-free medium combination. J. Neurosci. Res. 1993, 35, 567–576. [Google Scholar] [CrossRef] [PubMed]
- Bekkers, J.M.; Stevens, C.F. Presynaptic mechanism for long-term potentiation in the hippocampus. Nature 1990, 346, 724–729. [Google Scholar] [CrossRef] [PubMed]
- Goda, Y.; Stevens, C.F. Long-term depression properties in a simple system. Neuron 1996, 16, 103–111. [Google Scholar] [CrossRef]
- Bliss, T.V.; Lomo, T. Long-lasting potentiation of synaptic transmission in the dentate area of the anaesthetized rabbit following stimulation of the perforant path. J. Physiol. 1973, 232, 331–356. [Google Scholar] [CrossRef] [PubMed]
- Fitzsimonds, R.M.; Song, H.J.; Poo, M.M. Propagation of activity-dependent synaptic depression in simple neural networks. Nature 1997, 388, 439–448. [Google Scholar] [PubMed]
- Bi, G.; Poo, M. Distributed synaptic modification in neural networks induced by patterned stimulation. Nature 1999, 401, 792–796. [Google Scholar] [CrossRef] [PubMed]
- Bi, G.Q.; Poo, M.M. Synaptic modifications in cultured hippocampal neurons: Dependence on spike timing, synaptic strength, and postsynaptic cell type. J. Neurosci. Off. J. Soc. Neurosci. 1998, 18, 10464–10472. [Google Scholar]
- Wilcox, K.S.; Buchhalter, J.; Dichter, M.A. Properties of inhibitory and excitatory synapses between hippocampal neurons in very low density cultures. Synapse 1994, 18, 128–151. [Google Scholar] [CrossRef] [PubMed]
- Eytan, D.; Brenner, N.; Marom, S. Selective adaptation in networks of cortical neurons. J. Neurosci. Off. J. Soc. Neurosci. 2003, 23, 9349–9356. [Google Scholar]
- Chiappalone, M.; Massobrio, P.; Martinoia, S. Network plasticity in cortical assemblies. Eur. J. Neurosci. 2008, 28, 221–237. [Google Scholar] [CrossRef] [PubMed]
- Madhavan, R.; Chao, Z.C.; Potter, S.M. Plasticity of recurring spatiotemporal activity patterns in cortical networks. Phys. Biol. 2007, 4, 181–193. [Google Scholar] [CrossRef] [PubMed]
- Dranias, M.R.; Ju, H.; Rajaram, E.; VanDongen, A.M.J. Short-term memory in networks of dissociated cortical neurons. J. Neurosci. Off. J. Soc. Neurosci. 2013, 33, 1940–1953. [Google Scholar] [CrossRef] [PubMed]
- Chao, Z.C.; Bakkum, D.J.; Potter, S.M. Shaping embodied neural networks for adaptive goal-directed behavior. PLoS Comput. Biol. 2008, 4, e1000042. [Google Scholar] [CrossRef] [PubMed]
- Kleinfeld, D.; Kahler, K.H.; Hockberger, P.E. Controlled outgrowth of dissociated neurons on patterned substrates. J. Neurosci. Off. J. Soc. Neurosci. 1988, 8, 4098–4120. [Google Scholar]
- Qin, D.; Xia, Y.; Whitesides, G.M. Rapid prototyping of complex structures with feature sizes larger than 20 μm. Adv. Mater. 1996, 8, 917–919. [Google Scholar] [CrossRef]
- Duffy, D.C.; McDonald, J.C.; Schueller, O.J.; Whitesides, G.M. Rapid Prototyping of Microfluidic Systems in Poly(dimethylsiloxane). Anal. Chem. 1998, 70, 4974–4984. [Google Scholar] [CrossRef] [PubMed]
- Qin, D.; Xia, Y.; Whitesides, G.M. Soft lithography for micro- and nanoscale patterning. Nat. Protoc. 2010, 5, 491–502. [Google Scholar] [CrossRef] [PubMed]
- Singhvi, R.; Kumar, A.; Lopez, G.P.; Stephanopoulos, G.N.; Wang, D.I.; Whitesides, G.M.; Ingber, D.E. Engineering cell shape and function. Science 1994, 264, 696–698. [Google Scholar] [CrossRef] [PubMed]
- Branch, D.W.; Wheeler, B.C.; Brewer, G.J.; Leckband, D.E. Long-term maintenance of patterns of hippocampal pyramidal cells on substrates of polyethylene glycol and microstamped polylysine. IEEE Trans. Biomed. Eng. 2000, 47, 290–300. [Google Scholar] [CrossRef] [PubMed]
- Chang, J.C.; Brewer, G.J.; Wheeler, B.C. Modulation of neural network activity by patterning. Biosens. Bioelectron. 2001, 16, 527–533. [Google Scholar] [CrossRef]
- Marconi, E.; Nieus, T.; Maccione, A.; Valente, P.; Simi, A.; Messa, M.; Dante, S.; Baldelli, P.; Berdondini, L.; Benfenati, F. Emergent functional properties of neuronal networks with controlled topology. PLoS ONE 2012, 7, e34648. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Huang, Z.; Liu, W.; Yang, K.; Sun, K.; Xing, S.; Wang, D.; Zhang, W.; Jiang, X. Surface coating as a key parameter in engineering neuronal network structures in vitro. Biointerphases 2012, 7, 29. [Google Scholar] [CrossRef] [PubMed]
- Maher, M.P.; Pine, J.; Wright, J.; Tai, Y.C. The neurochip: A new multielectrode device for stimulating and recording from cultured neurons. J. Neurosci. Methods 1999, 87, 45–56. [Google Scholar] [CrossRef]
- Erickson, J.; Tooker, A.; Tai, Y.-C.; Pine, J. Caged neuron MEA: A system for long-term investigation of cultured neural network connectivity. J. Neurosci. Methods 2008, 175, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Xu, Z.; Huang, J.; Lin, X.; Luo, R.; Chen, C.-H.; Shi, P. NeuroArray: A universal interface for patterning and interrogating neural circuitry with single cell resolution. Sci. Rep. 2014, 4, 4784. [Google Scholar] [CrossRef] [PubMed]
- Taylor, A.M.; Blurton-Jones, M.; Rhee, S.W.; Cribbs, D.H.; Cotman, C.W.; Jeon, N.L. A microfluidic culture platform for CNS axonal injury, regeneration and transport. Nat. Methods 2005, 2, 599–605. [Google Scholar] [CrossRef] [PubMed]
- Wilson, N.R.; Ty, M.T.; Ingber, D.E.; Sur, M.; Liu, G. Synaptic reorganization in scaled networks of controlled size. J. Neurosci. Off. J. Soc. Neurosci. 2007, 27, 13581–13589. [Google Scholar] [CrossRef] [PubMed]
- Stenger, D.A.; Hickman, J.J.; Bateman, K.E.; Ravenscroft, M.S.; Ma, W.; Pancrazio, J.J.; Shaffer, K.; Schaffner, A.E.; Cribbs, D.H.; Cotman, C.W. Microlithographic determination of axonal/dendritic polarity in cultured hippocampal neurons. J. Neurosci. Methods 1998, 82, 167–173. [Google Scholar] [CrossRef]
- Edwards, D.; Stancescu, M.; Molnar, P.; Hickman, J.J. Two cell circuits of oriented adult hippocampal neurons on self-assembled monolayers for use in the study of neuronal communication in a defined system. ACS Chem. Neurosci. 2013, 4, 1174–1182. [Google Scholar] [CrossRef] [PubMed]
- Fricke, R.; Zentis, P.D.; Rajappa, L.T.; Hofmann, B.; Banzet, M.; Offenhäusser, A.; Meffert, S.H. Axon guidance of rat cortical neurons by microcontact printed gradients. Biomaterials 2011, 32, 2070–2076. [Google Scholar] [CrossRef] [PubMed]
- Tomba, C.; Braïni, C.; Wu, B.; Gov, N.S.; Villard, C. Tuning the adhesive geometry of neurons: Length and polarity control. Soft Matter 2014, 10, 2381–2387. [Google Scholar] [CrossRef] [PubMed]
- Claverol-Tinturé, E.; Ghirardi, M.; Fiumara, F.; Rosell, X.; Cabestany, J. Multielectrode arrays with elastomeric microstructured overlays for extracellular recordings from patterned neurons. J. Neural Eng. 2005, 2, L1–L7. [Google Scholar] [CrossRef] [PubMed]
- Claverol-Tinturé, E.; Cabestany, J.; Rosell, X. Multisite recording of extracellular potentials produced by microchannel-confined neurons in-vitro. IEEE Trans. Biomed. Eng. 2007, 54, 331–335. [Google Scholar] [CrossRef] [PubMed]
- Zhou, T.; Perry, S. F.; Berdichevsky, Y.; Petryna, S.; Fluck, V.; Tatic-Lucic, S. Multi-electrode array capable of supporting precisely patterned hippocampal neuronal networks. Biomed. Microdevices 2015, 17, 2. [Google Scholar] [CrossRef] [PubMed]
- Renault, R.; Sukenik, N.; Descroix, S.; Malaquin, L.; Viovy, J.-L.; Peyrin, J.-M.; Bottani, S.; Monceau, P.; Moses, E.; Vignes, M. Combining microfluidics, optogenetics and calcium imaging to study neuronal communication in vitro. PLoS ONE 2015, 10, e0120680. [Google Scholar] [CrossRef] [PubMed]
- Le Feber, J.; Postma, W.; de Weerd, E.; Weusthof, M.; Rutten, W.L.C. Barbed channels enhance unidirectional connectivity between neuronal networks cultured on multi electrode arrays. Front. Neurosci. 2015, 9, 412. [Google Scholar] [CrossRef] [PubMed]
- Pan, L.; Alagapan, S.; Franca, E.; Leondopulos, S.S.; DeMarse, T.B.; Brewer, G.J.; Wheeler, B.C. An in vitro method to manipulate the direction and functional strength between neural populations. Front. Neural Circuits 2015, 9, 32. [Google Scholar] [CrossRef] [PubMed]
- Deleglise, B.; Magnifico, S.; Duplus, E.; Vaur, P.; Soubeyre, V.; Belle, M.; Vignes, M.; Viovy, J.-L.; Jacotot, E.; Peyrin, J.-M.; et al. β-amyloid induces a dying-back process and remote trans-synaptic alterations in a microfluidic-based reconstructed neuronal network. Acta Neuropathol. Commun. 2014, 2, 145. [Google Scholar] [CrossRef] [PubMed]
- Tang-Schomer, M.D.; Davies, P.; Graziano, D.; Thurber, A.E.; Kaplan, D.L. Neural circuits with long-distance axon tracts for determining functional connectivity. J. Neurosci. Methods 2014, 222, 82–90. [Google Scholar] [CrossRef] [PubMed]
- Morin, F.; Nishimura, N.; Griscom, L.; Lepioufle, B.; Fujita, H.; Takamura, Y.; Tamiya, E. Constraining the connectivity of neuronal networks cultured on microelectrode arrays with microfluidic techniques: A step towards neuron-based functional chips. Biosens. Bioelectron. 2006, 21, 1093–1100. [Google Scholar] [CrossRef] [PubMed]
- DeMarse, T.B.; Pan, L.; Alagapan, S.; Brewer, G.J.; Wheeler, B.C. Feed-Forward Propagation of Temporal and Rate Information between Cortical Populations during Coherent Activation in Engineered In Vitro Networks. Front. Neural Circuits 2016, 10, 32. [Google Scholar] [CrossRef] [PubMed]
- Kanagasabapathi, T.T.; Massobrio, P.; Barone, R.A.; Tedesco, M.; Martinoia, S.; Wadman, W.J.; Decré, M.M.J. Functional connectivity and dynamics of cortical-thalamic networks co-cultured in a dual compartment device. J. Neural Eng. 2012, 9, 36010. [Google Scholar] [CrossRef] [PubMed]
- Peyrin, J.-M.; Deleglise, B.; Saias, L.; Vignes, M.; Gougis, P.; Magnifico, S.; Betuing, S.; Pietri, M.; Caboche, J.; Vanhoutte, P.; et al. Axon diodes for the reconstruction of oriented neuronal networks in microfluidic chambers. Lab Chip 2011, 11, 3663–3673. [Google Scholar] [CrossRef] [PubMed]
- Shi, P.; Scott, M.A.; Ghosh, B.; Wan, D.; Wissner-Gross, Z.; Mazitschek, R.; Haggarty, S.J.; Yanik, M.F. Synapse microarray identification of small molecules that enhance synaptogenesis. Nat. Commun. 2011, 2, 510. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Riss, M.; Buitrago, J.O.; Claverol-Tinturé, E. Biophysics of microchannel-enabled neuron-electrode interfaces. J. Neural Eng. 2012, 9, 26010. [Google Scholar] [CrossRef] [PubMed]
- Dworak, B.J.; Wheeler, B.C. Novel MEA platform with PDMS microtunnels enables the detection of action potential propagation from isolated axons in culture. Lab Chip 2009, 9, 404–410. [Google Scholar] [CrossRef] [PubMed]
- Shepherd, G.; Grillner, S. Handbook of Brain Microcircuits; Oxford University Press: Oxford, UK, 2010. [Google Scholar]
- Renault, R.; Durand, J.-B.; Viovy, J.-L.; Villard, C. Asymmetric axonal edge guidance: A new paradigm for building oriented neuronal networks. Lab Chip 2016, 16, 2188–2191. [Google Scholar] [CrossRef] [PubMed]
- Albers, J.; Offenhäusser, A. Signal Propagation between Neuronal Populations Controlled by Micropatterning. Front. Bioeng. Biotechnol. 2016, 4, 46. [Google Scholar] [CrossRef] [PubMed]
- Feinerman, O.; Rotem, A.; Moses, E. Reliable neuronal logic devices from patterned hippocampal cultures. Nat. Phys. 2008, 4, 967–973. [Google Scholar] [CrossRef]
- Edelman, D.B.; Keefer, E.W. A cultural renaissance: in vitro cell biology embraces three-dimensional context. Exp. Neurol. 2005, 192, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Gähwiler, B.H.; Capogna, M.; Debanne, D.; McKinney, R.A.; Thompson, S.M. Organotypic slice cultures: A technique has come of age. Trends Neurosci. 1997, 20, 471–477. [Google Scholar] [CrossRef]
- Gahwiler, B.; Thompson, S.; McKinney, A.; Debanne, D.; Robertson, R. Organotypic Slice Cultures of Neural Tissue. In Culturing Nerve Cells; Massachusetts Institute of Technology: Cambridge, MA, USA, 1998; pp. 461–498. [Google Scholar]
- Jahnsen, H.; Kristensen, B.W.; Thiébaud, P.; Noraberg, J.; Jakobsen, B.; Bove, M.; Martinoia, S.; Koudelka-Hep, M.; Grattarola, M.; Zimmer, J. Coupling of organotypic brain slice cultures to silicon-based arrays of electrodes. Methods 1999, 18, 160–172. [Google Scholar] [CrossRef] [PubMed]
- Kristensen, B.W.; Noraberg, J.; Thiébaud, P.; Koudelka-Hep, M.; Zimmer, J. Biocompatibility of silicon-based arrays of electrodes coupled to organotypic hippocampal brain slice cultures. Brain Res. 2001, 896, 1–17. [Google Scholar] [CrossRef]
- Van Bergen, A.; Papanikolaou, T.; Schuker, A.; Möller, A.; Schlosshauer, B. Long-term stimulation of mouse hippocampal slice culture on microelectrode array. Brain Res. Protoc. 2003, 11, 123–133. [Google Scholar] [CrossRef]
- Thiébaud, P.; de Rooij, N.F.; Koudelka-Hep, M.; Stoppini, L. Microelectrode arrays for electrophysiological monitoring of hippocampal organotypic slice cultures. IEEE Trans. Biomed. Eng. 1997, 44, 1159–1163. [Google Scholar] [CrossRef] [PubMed]
- Berdichevsky, Y.; Sabolek, H.; Levine, J.B.; Staley, K.J.; Yarmush, M.L. Microfluidics and multielectrode array-compatible organotypic slice culture method. J. Neurosci. Methods 2009, 178, 59–64. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Pan, L.; Cheng, X.; Berdichevsky, Y. Perfused drop microfluidic device for brain slice culture-based drug discovery. Biomed. Microdevices 2016, 18, 46. [Google Scholar] [CrossRef] [PubMed]
- Berdichevsky, Y.; Staley, K.J.; Yarmush, M.L. Building and manipulating neural pathways with microfluidics. Lab Chip 2010, 10, 999–1004. [Google Scholar] [CrossRef] [PubMed]
- Dana, H.; Marom, A.; Paluch, S.; Dvorkin, R.; Brosh, I.; Shoham, S. Hybrid multiphoton volumetric functional imaging of large-scale bioengineered neuronal networks. Nat. Commun. 2014, 5, 3997. [Google Scholar] [CrossRef] [PubMed]
- Odawara, A.; Gotoh, M.; Suzuki, I. Control of neural network patterning using collagen gel photothermal etching. Lab Chip 2013, 13, 2040–2046. [Google Scholar] [CrossRef] [PubMed]
- Honegger, T.; Thielen, M.I.; Feizi, S.; Sanjana, N.E.; Voldman, J. Microfluidic neurite guidance to study structure-function relationships in topologically-complex population-based neural networks. Sci. Rep. 2016, 6, 28384. [Google Scholar] [CrossRef] [PubMed]
- Pautot, S.; Wyart, C.; Isacoff, E.Y. Colloid-guided assembly of oriented 3D neuronal networks. Nat. Methods 2008, 5, 735–740. [Google Scholar] [CrossRef] [PubMed]
- Schüz, A.; Palm, G. Density of neurons and synapses in the cerebral cortex of the mouse. J. Comp. Neurol. 1989, 286, 442–455. [Google Scholar] [CrossRef] [PubMed]
- Frega, M.; Tedesco, M.; Massobrio, P.; Pesce, M.; Martinoia, S. Network dynamics of 3D engineered neuronal cultures: A new experimental model for in-vitro electrophysiology. Sci. Rep. 2014, 4, 5489. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.; Sun, Y.; Liu, W.; Zhang, W.; Zheng, W.; Jiang, X. Assembly of functional three-dimensional neuronal networks on a microchip. Small 2014, 10, 2530–2536. [Google Scholar] [CrossRef] [PubMed]
- Xie, J.; Liu, W.; MacEwan, M.R.; Yeh, Y.-C.; Thomopoulos, S.; Xia, Y. Nanofiber membranes with controllable microwells and structural cues and their use in forming cell microarrays and neuronal networks. Small 2011, 7, 293–297. [Google Scholar] [CrossRef] [PubMed]
- Kato-Negishi, M.; Morimoto, Y.; Onoe, H.; Takeuchi, S. Millimeter-sized neural building blocks for 3D heterogeneous neural network assembly. Adv. Healthc. Mater. 2013, 2, 1564–1570. [Google Scholar] [CrossRef] [PubMed]
- Hodgkin, A.L.; Huxley, A.F. Action potentials recorded from inside a nerve fibre. Nature 1939, 144, 710–711. [Google Scholar] [CrossRef]
- Verkhratsky, A.; Krishtal, O.A.; Petersen, O.H. From Galvani to patch clamp: the development of electrophysiology. Pflüg Arch. Eur. J. Physiol. 2006, 453, 233–247. [Google Scholar] [CrossRef] [PubMed]
- Blanton, M.G.; Lo Turco, J.J.; Kriegstein, A.R. Whole cell recording from neurons in slices of reptilian and mammalian cerebral cortex. J. Neurosci. Methods 1989, 30, 203–210. [Google Scholar] [CrossRef]
- Spira, M.E.; Hai, A. Multi-electrode array technologies for neuroscience and cardiology. Nat. Nanotechnol. 2013, 8, 83–94. [Google Scholar] [CrossRef] [PubMed]
- Henze, D.A.; Borhegyi, Z.; Csicsvari, J.; Mamiya, A.; Harris, K.D.; Buzsáki, G. Intracellular features predicted by extracellular recordings in the hippocampus in vivo. J. Neurophysiol. 2000, 84, 390–400. [Google Scholar] [PubMed]
- Harris, K.D.; Henze, D.A.; Csicsvari, J.; Hirase, H.; Buzsáki, G. Accuracy of tetrode spike separation as determined by simultaneous intracellular and extracellular measurements. J. Neurophysiol. 2000, 84, 401–414. [Google Scholar] [PubMed]
- Maccione, A.; Gandolfo, M.; Zordan, S.; Amin, H.; Di Marco, S.; Nieus, T.; Angotzi, G.N.; Berdondini, L. Microelectronics, bioinformatics and neurocomputation for massive neuronal recordings in brain circuits with large scale multielectrode array probes. Brain Res. Bull. 2015, 119, 118–126. [Google Scholar] [CrossRef] [PubMed]
- Smetters, D.; Majewska, A.; Yuste, R. Detecting action potentials in neuronal populations with calcium imaging. Methods 1999, 18, 215–221. [Google Scholar] [CrossRef] [PubMed]
- Grynkiewicz, G.; Poenie, M.; Tsien, R.Y. A new generation of Ca2+ indicators with greatly improved fluorescence properties. J. Biol. Chem. 1985, 260, 3440–3450. [Google Scholar] [PubMed]
- Chen, T.-W.; Wardill, T.J.; Sun, Y.; Pulver, S.R.; Renninger, S.L.; Baohan, A.; Schreiter, E.R.; Kerr, R.A.; Orger, M.B.; Jayaraman, V.; et al. Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature 2013, 499, 295–300. [Google Scholar] [CrossRef] [PubMed]
- Helmchen, F.; Imoto, K.; Sakmann, B. Ca2+ buffering and action potential-evoked Ca2+ signaling in dendrites of pyramidal neurons. Biophys. J. 1996, 70, 1069–1081. [Google Scholar] [CrossRef]
- Jin, L.; Han, Z.; Platisa, J.; Wooltorton, J.R.A.; Cohen, L.B.; Pieribone, V.A. Single action potentials and subthreshold electrical events imaged in neurons with a fluorescent protein voltage probe. Neuron 2012, 75, 779–785. [Google Scholar] [CrossRef] [PubMed]
- St-Pierre, F.; Chavarha, M.; Lin, M.Z. Designs and sensing mechanisms of genetically encoded fluorescent voltage indicators. Curr. Opin. Chem. Biol. 2015, 27, 31–38. [Google Scholar] [CrossRef] [PubMed]
- Gong, Y.; Huang, C.; Li, J.Z.; Grewe, B.F.; Zhang, Y.; Eismann, S.; Schnitzer, M.J. High-speed recording of neural spikes in awake mice and flies with a fluorescent voltage sensor. Science 2015, 350, 1361–1366. [Google Scholar] [CrossRef] [PubMed]
- Nagel, G.; Szellas, T.; Huhn, W.; Kateriya, S.; Adeishvili, N.; Berthold, P.; Ollig, D.; Hegemann, P.; Bamberg, E. Channelrhodopsin-2, a directly light-gated cation-selective membrane channel. Proc. Natl. Acad. Sci. USA 2003, 100, 13940–13945. [Google Scholar] [CrossRef] [PubMed]
- Boyden, E.S.; Zhang, F.; Bamberg, E.; Nagel, G.; Deisseroth, K. Millisecond-timescale, genetically targeted optical control of neural activity. Nat. Neurosci. 2005, 8, 1263–1268. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Araki, S.; Wu, J.; Teramoto, T.; Chang, Y.-F.; Nakano, M.; Abdelfattah, A.S.; Fujiwara, M.; Ishihara, T.; Nagai, T.; et al. An expanded palette of genetically encoded Ca2+ indicators. Science 2011, 333, 1888–1891. [Google Scholar] [CrossRef] [PubMed]
- Akerboom, J.; Carreras Calderón, N.; Tian, L.; Wabnig, S.; Prigge, M.; Tolö, J.; Gordus, A.; Orger, M.B.; Severi, K.E.; Macklin, J.J.; et al. Genetically encoded calcium indicators for multi-color neural activity imaging and combination with optogenetics. Front. Mol. Neurosci. 2013, 6, 2. [Google Scholar] [CrossRef] [PubMed]
- Yizhar, O.; Fenno, L.E.; Davidson, T.J.; Mogri, M.; Deisseroth, K. Optogenetics in neural systems. Neuron 2011, 71, 9–34. [Google Scholar] [CrossRef] [PubMed]
- Turrigiano, G.G.; Leslie, K.R.; Desai, N.S.; Rutherford, L.C.; Nelson, S.B. Activity-dependent scaling of quantal amplitude in neocortical neurons. Nature 1998, 391, 892–896. [Google Scholar] [CrossRef] [PubMed]
- Markram, H.; Lübke, J.; Frotscher, M.; Sakmann, B. Regulation of synaptic efficacy by coincidence of postsynaptic APs and EPSPs. Science 1997, 275, 213–215. [Google Scholar] [CrossRef] [PubMed]
- Wolf, F.; Geisel, T. Logic gates come to life. Nat. Phys. 2008, 4, 905–906. [Google Scholar] [CrossRef]
- Marblestone, A.H.; Zamft, B.M.; Maguire, Y.G.; Shapiro, M.G.; Cybulski, T.R.; Glaser, J.I.; Amodei, D.; Stranges, P.B.; Kalhor, R.; Dalrymple, D.A.; et al. Physical principles for scalable neural recording. Front. Comput. Neurosci. 2013, 7, 137. [Google Scholar] [CrossRef] [PubMed]





© 2016 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hasan, M.F.; Berdichevsky, Y. Neural Circuits on a Chip. Micromachines 2016, 7, 157. https://doi.org/10.3390/mi7090157
Hasan MF, Berdichevsky Y. Neural Circuits on a Chip. Micromachines. 2016; 7(9):157. https://doi.org/10.3390/mi7090157
Chicago/Turabian StyleHasan, Md. Fayad, and Yevgeny Berdichevsky. 2016. "Neural Circuits on a Chip" Micromachines 7, no. 9: 157. https://doi.org/10.3390/mi7090157
APA StyleHasan, M. F., & Berdichevsky, Y. (2016). Neural Circuits on a Chip. Micromachines, 7(9), 157. https://doi.org/10.3390/mi7090157