Classification of Breast Cancer Cells Using the Integration of High-Frequency Single-Beam Acoustic Tweezers and Convolutional Neural Networks
Abstract
:1. Introduction
2. Results
3. Discussion
4. Material and Method
4.1. Transducer Fabrication
4.2. Transducer Performance
4.3. Cell Preparation
4.4. Live Intracellular Calcium Imaging
4.5. SBAT for Cell Deformation
4.6. Cancer Cell Classification with Convolutional Neural Networks
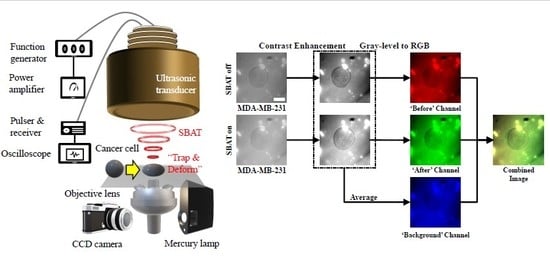
4.6.1. Preprocessing
- Enhance contrast of cell images.
- Put the SBAT on images as the red channel, the SBAT off images as the green channel, and the average of the SBAT on and off images as the blue channel.
- Save the combined image.
4.6.2. CNN Model for Cancer Cell Classification
4.7. Cell Viability Test
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Zhang, N.; Wu, J.; Yang, M.; Yu, J.; Li, R. A Composite Model Integrating Imaging, Histological, and Genetic Features to Predict Tumor Mutation Burden in Non-Small Cell Lung Cancer Patients. Int. J. Radiat. Oncol. Biol. Phys. 2019, 105, E541. [Google Scholar] [CrossRef]
- Rossi, G.; Berbellini, A.; Ancidei, S.; Montini, G.C.; Capoccetti, F.; Cidda, C.; Fattori, S.; Cardarelli, M.; Patrizi, I.; Brianzoni, E. Calculation of Lesion Volume in Non-Small Cell Lung Cancer (NSCLC) by PET/CT Imaging: Histological Comparation and Threshold Study. Preliminary Report. Eur. J. Nucl. Med. Mol. Imaging 2015, 32, S37. [Google Scholar]
- Paschali, A.; Papandrianos, N.; Koletsis, E.; Stamou, E.; Spyridonidis, T.; Savvopoulos, C.; Barla, P.; Dougenis, D.; Vassilakos, P.J.; Apostolopoulos, D.J. The Value Of 99mTc-Depreotide SPECT/CT Imaging For Lymph Node Staging In Non-Small-Cell Lung Cancer. A Study With Histological Confirmation. Eur. J. Nucl. Med. Mol. Imaging 2009, 36, S361–S362. [Google Scholar]
- Gallastegui, A.; Cheung, J.; Southard, T.; Hume, K.R. Volumetric and linear measurements of lung tumor burden from non-gated micro-CT imaging correlate with histological analysis in a genetically engineered mouse model of non-small cell lung cancer. Lab. Anim. 2018, 52, 457–469. [Google Scholar] [CrossRef] [PubMed]
- Kurc, T.; Sharma, A.; Oh, T.; Farris, A.; Saltz, J.; Wang, F.; Cooper, L.; Kong, J.; Pan, T.; Chen, W.; et al. A data model and database for high-resolution pathology analytical image informatics. J. Pathol. Inform. 2011, 2, 32. [Google Scholar] [CrossRef] [PubMed]
- Nelissen, B.G.L.; van Herwaarden, J.A.; Moll, F.L.; van Diest, P.J.; Pasterkamp, G. SlideToolkit: An Assistive Toolset for the Histological Quantification of Whole Slide Images. PLoS ONE 2014, 9, e110289. [Google Scholar] [CrossRef]
- Zerbe, N.; Hufnagl, P.; Schlüns, K. Distributed computing in image analysis using open source frameworks and application to image sharpness assessment of histological whole slide images. Diagn. Pathol. 2011, 6, S16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Q.; Lee, G.; Ong, C.; Lim, C. AFM indentation study of breast cancer cells. Biochem. Biophys. Res. Commun. 2008, 374, 609–613. [Google Scholar] [CrossRef]
- Cross, S.E.; Jin, Y.S.; Tondre, J.; Wong, R.; Rao, J.; Gimzewski, J.K. AFM-based analysis of human metastatic cancer cells. Nanotechnology 2008, 19, 384003. [Google Scholar] [CrossRef]
- Rico, F.; Roca-Cusachs, P.; Gavara, N.; Farré, R.; Rotger, M.; Navajas, D. Probing mechanical properties of living cells by atomic force microscopy with blunted pyramidal cantilever tips. Phys. Rev. E 2005, 72, 021914. [Google Scholar] [CrossRef] [Green Version]
- Coceano, G.; Yousafzai, M.S.; Ma, W.; Ndoye, F.; Venturelli, L.; Hussain, I.; Bonin, S.; Niemela, J.; Scoles, G.; Cojoc, D.; et al. Investigation into local cell mechanics by atomic force microscopy mapping and optical tweezer vertical indentation. Nanotechnology 2015, 27, 065102. [Google Scholar] [CrossRef] [PubMed]
- Grier, D.G. A revolution in optical manipulation. Nature 2003, 424, 810–816. [Google Scholar] [CrossRef] [PubMed]
- Walter, N.; Selhuber, C.; Kessler, H.; Spatz, J.P. Cellular Unbinding Forces of Initial Adhesion Processes on Nanopatterned Surfaces Probed with Magnetic Tweezers. Nano Lett. 2006, 6, 398–402. [Google Scholar] [CrossRef]
- Yoon, J.K.; Lee, T.I.; Bhang, S.H.; Shin, J.Y.; Myoung, J.M.; Kim, B.S. Stretchable Piezoelectric Substrate Providing Pulsatile Mechanoelectric Cues for Cardiomyogenic Differentiation of Mesenchymal Stem Cells. ACS Appl. Mater. Interfaces 2017, 9, 22101–22111. [Google Scholar] [CrossRef]
- Melzer, J.E.; McLeod, E. Fundamental Limits of Optical Tweezer Nanoparticle Manipulation Speeds. ACS Nano 2018, 12, 2440–2447. [Google Scholar] [CrossRef] [PubMed]
- Lim, H.G.; Li, Y.; Lin, M.Y.; Yoon, C.; Lee, C.; Jung, H.; Chow, R.H.; Shung, K.K. Calibration of Trapping Force on Cell-Size Objects From Ultrahigh-Frequency Single-Beam Acoustic Tweezer. IEEE Trans. Ultrason. Ferroelectr. Freq. Control 2016, 63, 1988–1995. [Google Scholar] [CrossRef]
- Lim, H.G.; Kim, H.H.; Yoon, C. Evaluation method for acoustic trapping performance by tracking motion of trapped microparticle. Jpn. J. Appl. Phys. 2018, 57, 057202. [Google Scholar] [CrossRef]
- Lim, H.G.; Kim, H.H.; Yoon, C.; Shung, K.K. A One-Sided Acoustic Trap for Cell Immobilization Using 30-MHz Array Transducer. IEEE Trans. Ultrason. Ferroelectr. Freq. Control 2020, 67, 167–172. [Google Scholar] [CrossRef]
- Lam, K.H.; Li, Y.; Li, Y.; Lim, H.G.; Zhou, Q.; Shung, K.K. Multifunctional single beam acoustic tweezer for non-invasive cell/organism manipulation and tissue imaging. Sci. Rep. 2016, 6. [Google Scholar] [CrossRef] [Green Version]
- Hwang, J.Y.; Yoon, C.W.; Lim, H.G.; Park, J.M.; Yoon, S.; Lee, J.; Shung, K.K. Acoustic tweezers for studying intracellular calcium signaling in SKBR-3 human breast cancer cells. Ultrasonics 2015, 63, 94–101. [Google Scholar] [CrossRef] [Green Version]
- Lim, H.G.; Shung, K.K. Quantification of Inter-Erythrocyte Forces with Ultra-High Frequency (410 MHz) Single Beam Acoustic Tweezer. Ann. Biomed. Eng. 2017, 45, 2174–2183. [Google Scholar] [CrossRef]
- Liu, H.C.; Gang, E.J.; Kim, H.N.; Lim, H.G.; Jung, H.; Chen, R.; Abdel-Azim, H.; Shung, K.K.; Kim, Y.M. Characterizing Deformability of Drug Resistant Patient-Derived Acute Lymphoblastic Leukemia (ALL) Cells Using Acoustic Tweezers. Sci. Rep. 2018, 8. [Google Scholar] [CrossRef]
- Hwang, J.Y.; Kim, J.; Park, J.M.; Lee, C.; Jung, H.; Lee, J.; Shung, K.K. Cell Deformation by Single-beam Acoustic Trapping: A Promising Tool for Measurements of Cell Mechanics. Sci. Rep. 2016, 6. [Google Scholar] [CrossRef]
- Lee, J.; Lee, C.; Shung, K.K. Calibration of sound forces in acoustic traps. IEEE Trans. Ultrason. Ferroelectr. Freq. Control 2010, 57, 2305–2310. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.; Jeong, J.S.; Shung, K.K. Microfluidic acoustic trapping force and stiffness measurement using viscous drag effect. Ultrasonics 2013, 53, 249–254. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Y.; Lee, C.; Lam, K.H.; Shung, K.K. A simple method for evaluating the trapping performance of acoustic tweezers. Appl. Phys. Lett. 2013, 102, 084102. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hwang, J.Y.; Lim, H.G.; Yoon, C.W.; Lam, K.H.; Yoon, S.; Lee, C.; Chiu, C.T.; Kang, B.J.; Kim, H.H.; Shung, K.K. Non-contact High-Frequency Ultrasound Microbeam Stimulation for Studying Mechanotransduction in Human Umbilical Vein Endothelial Cells. Ultrasound Med. Biol. 2014, 40, 2172–2182. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, M.G.; Park, J.; Lim, H.G.; Yoon, S.; Lee, C.; Chang, J.H.; Shung, K.K. Label-free analysis of the characteristics of a single cell trapped by acoustic tweezers. Sci. Rep. 2017, 7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hwang, J.Y.; Lee, C.; Lam, K.H.; Kim, H.H.; Lee, J.; Shung, K.K. Cell membrane deformation induced by a fibronectin-coated polystyrene microbead in a 200-MHz acoustic trap. IEEE Trans. Ultrason. Ferroelectr. Freq. Control 2014, 61, 399–406. [Google Scholar] [CrossRef] [Green Version]
- Calzado-Martín, A.; Encinar, M.; Tamayo, J.; Calleja, M.; Paulo, A.S. Effect of Actin Organization on the Stiffness of Living Breast Cancer Cells Revealed by Peak-Force Modulation Atomic Force Microscopy. ACS Nano 2016, 10, 3365–3374. [Google Scholar] [CrossRef] [Green Version]
- Nikkhah, M.; Strobl, J.S.; Schmelz, E.M.; Agah, M. Evaluation of the influence of growth medium composition on cell elasticity. J. Biomech. 2011, 44, 762–766. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Guo, P.; Auguste, D.T. Mapping the CXCR4 receptor on breast cancer cells. Biomaterials 2015, 57, 161–168. [Google Scholar] [CrossRef] [PubMed]
- Cross, S.E.; Jin, Y.S.; Rao, J.; Gimzewski, J.K. Nanomechanical analysis of cells from cancer patients. Nat. Nanotechnol. 2007, 2, 780–783. [Google Scholar] [CrossRef] [PubMed]
- Darling, E.M.; Topel, M.; Zauscher, S.; Vail, T.P.; Guilak, F. Viscoelastic properties of human mesenchymally-derived stem cells and primary osteoblasts, chondrocytes, and adipocytes. J. Biomech. 2008, 41, 454–464. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Faria, E.C.; Ma, N.; Gazi, E.; Gardner, P.; Brown, M.; Clarke, N.; Snook, R.D. Measurement of elastic properties of prostate cancer cells using AFM. Anal. 2008, 133, 1498–1500. [Google Scholar] [CrossRef] [PubMed]
- Ruder, S. An Overview of Gradient Descent Optimization Algorithms. arXiv 2016, arXiv:1609.04747. [Google Scholar]
- Duchi, J.; Hazan, E.; Singer, Y. Adaptive Subgradient Methods for Online Learning and Stochastic Optimization. J. Mach. Learn. Res. 2011, 12, 2121–2159. [Google Scholar]
- Zeiler, M.D. ADADELTA: An Adaptive Learning Rate Method. arXiv 2012, arXiv:1212.5701. [Google Scholar]
- Kingma, D.P.; Ba, J. Adam: A Method for Stochastic Optimization. arXiv 2014, arXiv:1412.6980. [Google Scholar]
- Youn, S.; Choi, J.W.; Lee, J.S.; Kim, J.; Yang, I.H.; Chang, J.H.; Kim, H.C.; Hwang, J.Y. Acoustic Trapping Technique for Studying Calcium Response of a Suspended Breast Cancer Cell: Determination of Its Invasion Potentials. IEEE Trans. Ultrason. Ferroelectr. Freq. Control 2019, 66, 737–746. [Google Scholar] [CrossRef]
- Hwang, J.Y.; Lee, N.S.; Lee, C.; Lam, K.H.; Kim, H.H.; Woo, J.; Lin, M.Y.; Kisler, K.; Choi, H.; Zhou, Q.; et al. Investigating contactless high frequency ultrasound microbeam stimulation for determination of invasion potential of breast cancer cells. Biotechnol. Bioeng. 2013, 110, 2697–2705. [Google Scholar] [CrossRef] [PubMed] [Green Version]










| Parameter | Range | Step Size |
|---|---|---|
| Learning rate () | ||
| Batch size () | ||
| Epochs () |
| Optimizer | Validating Loss | Accuracy | Precision | Recall | Measure |
|---|---|---|---|---|---|
| SDG | 0.25 ** (0.20 *) | 0.91 (0.06) | 0.92 (0.07 *) | 0.91 (0.08) | 0.90 (0.07) |
| RMSprop | 0.89 (1.65) | 0.96 ** (0.05 **) | 0.93 ** (0.08) | 0.99 * (0.01 *) | 0.95 ** (0.05 **) |
| Adagrad | 0.21 * (0.25 **) | 0.85 (0.19) | 0.83 (0.19) | 0.98 (0.02) | 0.88 (0.13) |
| Adadelta | 1.51 (2.94) | 0.97 * (0.05 *) | 0.96 * (0.08 **) | 0.99 ** (0.01 **) | 0.97 * (0.05 *) |
| Adam | 0.57 (0.71) | 0.88 (0.09) | 0.86 (0.13) | 0.93 (0.07) | 0.88 (0.09) |
| Optimizer | Validating Loss | Accuracy | Precision | Recall | Measure |
|---|---|---|---|---|---|
| RMSprop | 0.00 | 0.90 | 0.97 | 0.84 | 0.89 |
| Adadelta | 8.49 | 0.85 | 0.87 | 0.82 | 0.82 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lim, H.G.; Lee, O.-J.; Shung, K.K.; Kim, J.-T.; Kim, H.H. Classification of Breast Cancer Cells Using the Integration of High-Frequency Single-Beam Acoustic Tweezers and Convolutional Neural Networks. Cancers 2020, 12, 1212. https://doi.org/10.3390/cancers12051212
Lim HG, Lee O-J, Shung KK, Kim J-T, Kim HH. Classification of Breast Cancer Cells Using the Integration of High-Frequency Single-Beam Acoustic Tweezers and Convolutional Neural Networks. Cancers. 2020; 12(5):1212. https://doi.org/10.3390/cancers12051212
Chicago/Turabian StyleLim, Hae Gyun, O-Joun Lee, K. Kirk Shung, Jin-Taek Kim, and Hyung Ham Kim. 2020. "Classification of Breast Cancer Cells Using the Integration of High-Frequency Single-Beam Acoustic Tweezers and Convolutional Neural Networks" Cancers 12, no. 5: 1212. https://doi.org/10.3390/cancers12051212
APA StyleLim, H. G., Lee, O. -J., Shung, K. K., Kim, J. -T., & Kim, H. H. (2020). Classification of Breast Cancer Cells Using the Integration of High-Frequency Single-Beam Acoustic Tweezers and Convolutional Neural Networks. Cancers, 12(5), 1212. https://doi.org/10.3390/cancers12051212