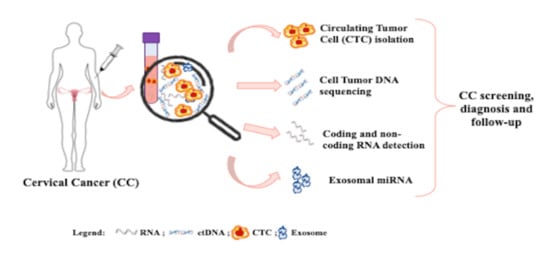
Liquid Biopsy in Cervical Cancer: Hopes and Pitfalls
Abstract
:Simple Summary
Abstract
1. Introduction
2. Circulating Tumor Cells (CTCs)
3. Circulating Cell-Free DNA (ccfDNA)
4. Total and Cell-Free Circulating RNA
4.1. Coding RNAs
4.2. Non-Coding RNAs
| Type of RNA | Biomarker Name | N. of CC Patients | Putative Clinical Validity | References |
|---|---|---|---|---|
| mRNA | HPV-16 E6 mRNA HPV-18 E6 mRNA | 35 | Prognostic | [59] |
| mRNA | EGFR mRNA | 45 | Prognostic | [60] |
| mRNA | Bmi-1 mRNA | 109 | Diagnostic Prognostic | [61] |
| lncRNAs | HOTAIR, PVT1, AL592284.1, XLOC_000303 | 300 | Diagnostic | [66] |
| lncRNA | AC017078.1 XLOC_011152 | 24 | Diagnostic | [67] |
| lncRNA | lncRNA DLX6-AS1 | 114 | Diagnostic Prognostic | [68] |
| miRNA | miR-218 | 90 | Diagnostic Prognostic | [69] |
| miRNA | miR-20a | 80 | Diagnostic | [70] |
| miRNAs | miR-20a, miR-1246, miR-2392, miR-3147, miR-3162-5p, miR-4484 | 80 | Prognostic | [71] |
| miRNA | miR-196a | 105 | Diagnostic Prognostic | [72] |
| miRNAs | miR-21, miR-25, miR-29a, miR-200a, miR-486-5p | 213 | Diagnostic | [73] |
| miRNA | miR-138 | Pre-clinical study | Therapeutic | [74] |
| miRNA | miR-148b | Pre-clinical study | Therapeutic | [75] |
| miRNA | miR-425-5p | 40 | Diagnostic Prognostic | [76] |
| miRNA | miR-30e | Pre-clinical study | Therapeutic | [77] |
| miRNA | miR-187 | 60 | Prognostic | [78] |
| Pre-clinical study | Therapeutic | |||
| miRNA | miR-138 | 168 | Prognostic | [79] |
| Pre-clinical study | Therapeutic | |||
| miRNA | miR-195 | Pre-clinical study | Therapeutic | [80] |
| miRNA | miR-214 | Pre-clinical study | Therapeutic | [81] |
| miRNA | miR-486-5p | Pre-clinical study | Diagnostic Therapeutic | [82] |
| miRNAs | miR-17-5p, miR-32-5p, miR-409-3p, miR-454-3p | 115 | Diagnostic | [83] |
| miRNAs + protein | miR-25, -29a, -486-5p (+ SCC Ag) | 200 | Diagnostic | [84] |
| exosomal miRNAs | let-7d-3p, miR-30d-5p | 63 | Diagnostic | [85] |
| exosomal miRNA | miR-125a-5p | 44 | Diagnostic Prognostic | [86] |
5. Exosomal miRNAs
6. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Lowy, D.R.; Solomon, D.; Hildesheim, A.; Schiller, J.T.; Schiffman, M. Human papillomavirus infection and the primary and secondary prevention of cervical cancer. Cancer 2008, 113, 1980–1993. [Google Scholar] [CrossRef]
- Arbyn, M.; Weiderpass, E.; Bruni, L.; de Sanjose, S.; Saraiya, M.; Ferlay, J.; Bray, F. Estimates of incidence and mortality of cervical cancer in 2018: A worldwide analysis. Lancet Glob. Health 2020, 8, e191–e203. [Google Scholar] [CrossRef] [Green Version]
- Marth, C.; Landoni, F.; Mahner, S.; McCormack, M.; Gonzalez-Martin, A.; Colombo, N.; Committee, E.G. Cervical cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann. Oncol. 2017, 28, iv72–iv83. [Google Scholar] [CrossRef]
- Chesson, H.W.; Dunne, E.F.; Hariri, S.; Markowitz, L.E. The estimated lifetime probability of acquiring human papillomavirus in the United States. Sex. Transm. Dis. 2014, 41, 660–664. [Google Scholar] [CrossRef] [PubMed]
- Gates, A.; Pillay, J.; Reynolds, D.; Stirling, R.; Traversy, G.; Korownyk, C.; Moore, A.; Theriault, G.; Thombs, B.D.; Little, J.; et al. Screening for the prevention and early detection of cervical cancer: Protocol for systematic reviews to inform Canadian recommendations. Syst. Rev. 2021, 10, 2. [Google Scholar] [CrossRef]
- Liang, L.A.; Einzmann, T.; Franzen, A.; Schwarzer, K.; Schauberger, G.; Schriefer, D.; Radde, K.; Zeissig, S.R.; Ikenberg, H.; Meijer, C.; et al. Cervical Cancer Screening: Comparison of Conventional Pap Smear Test, Liquid-Based Cytology, and Human Papillomavirus Testing as Stand-alone or Cotesting Strategies. Cancer Epidemiol. Biomarkers Prev. 2021, 30, 474–484. [Google Scholar] [CrossRef] [PubMed]
- Ronco, G.; Dillner, J.; Elfstrom, K.M.; Tunesi, S.; Snijders, P.J.; Arbyn, M.; Kitchener, H.; Segnan, N.; Gilham, C.; Giorgi-Rossi, P.; et al. Efficacy of HPV-based screening for prevention of invasive cervical cancer: Follow-up of four European randomised controlled trials. Lancet 2014, 383, 524–532. [Google Scholar] [CrossRef]
- Aniebue UU, O.T. Ethical, Socioeconomic, and Cultural Considerations in Gynecologic Cancer Care in Developing Countries". nt. J. Palliat. Care 2014, 2014, 6. [Google Scholar] [CrossRef] [Green Version]
- Isa Modibbo, F.; Dareng, E.; Bamisaye, P.; Jedy-Agba, E.; Adewole, A.; Oyeneyin, L.; Olaniyan, O.; Adebamowo, C. Qualitative study of barriers to cervical cancer screening among Nigerian women. BMJ Open 2016, 6, e008533. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bayrami, R.; Taghipour, A.; Ebrahimipour, H. Personal and socio-cultural barriers to cervical cancer screening in Iran, patient and provider perceptions: A qualitative study. Asian Pac. J. Cancer Prev. 2015, 16, 3729–3734. [Google Scholar] [CrossRef]
- Devarapalli, P.; Labani, S.; Nagarjuna, N.; Panchal, P.; Asthana, S. Barriers affecting uptake of cervical cancer screening in low and middle income countries: A systematic review. Indian J. Cancer 2018, 55, 318–326. [Google Scholar] [CrossRef] [PubMed]
- Vahabi, M.; Lofters, A. Muslim immigrant women’s views on cervical cancer screening and HPV self-sampling in Ontario, Canada. BMC Public Health 2016, 16, 868. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Palmirotta, R.; Silvestris, E.; D’Oronzo, S.; Cardascia, A.; Silvestris, F. Ovarian cancer: Novel molecular aspects for clinical assessment. Crit. Rev. Oncol. Hematol. 2017, 117, 12–29. [Google Scholar] [CrossRef]
- Liu, K.; Tong, H.; Li, T.; Chen, Y.; Mao, X. Potential value of circulating tumor DNA in gynecological tumors. Am. J. Transl. Res. 2020, 12, 3225–3233. [Google Scholar] [PubMed]
- Palmirotta, R.; Lovero, D.; Silvestris, E.; Felici, C.; Quaresmini, D.; Cafforio, P.; Silvestris, F. Next-generation Sequencing (NGS) Analysis on Single Circulating Tumor Cells (CTCs) with No Need of Whole-genome Amplification (WGA). Cancer Genom. Proteom. 2017, 14, 173–179. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Palmirotta, R.; Lovero, D.; Cafforio, P.; Felici, C.; Mannavola, F.; Pelle, E.; Quaresmini, D.; Tucci, M.; Silvestris, F. Liquid biopsy of cancer: A multimodal diagnostic tool in clinical oncology. Ther. Adv. Med. Oncol. 2018, 10, 1758835918794630. [Google Scholar] [CrossRef]
- Leon, S.A.; Shapiro, B.; Sklaroff, D.M.; Yaros, M.J. Free DNA in the serum of cancer patients and the effect of therapy. Cancer Res. 1977, 37, 646–650. [Google Scholar]
- Neumann, M.H.D.; Bender, S.; Krahn, T.; Schlange, T. ctDNA and CTCs in Liquid Biopsy—Current Status and Where We Need to Progress. Comput. Struct. Biotechnol. J. 2018, 16, 190–195. [Google Scholar] [CrossRef]
- Tellez-Gabriel, M.; Knutsen, E.; Perander, M. Current Status of Circulating Tumor Cells, Circulating Tumor DNA, and Exosomes in Breast Cancer Liquid Biopsies. Int. J. Mol. Sci. 2020, 21, 9457. [Google Scholar] [CrossRef]
- Vasseur, A.; Kiavue, N.; Bidard, F.C.; Pierga, J.Y.; Cabel, L. Clinical utility of circulating tumor cells: An update. Mol. Oncol. 2021, 15, 1647–1666. [Google Scholar] [CrossRef]
- Hench, I.B.; Hench, J.; Tolnay, M. Liquid Biopsy in Clinical Management of Breast, Lung, and Colorectal Cancer. Front. Med. 2018, 5, 9. [Google Scholar] [CrossRef]
- Tucci, M.; D’Oronzo, S.; Mannavola, F.; Felici, C.; Lovero, D.; Cafforio, P.; Palmirotta, R.; Silvestris, F. Dual-procedural separation of CTCs in cutaneous melanoma provides useful information for both molecular diagnosis and prognosis. Ther. Adv. Med. Oncol. 2020, 12, 1758835920905415. [Google Scholar] [CrossRef] [Green Version]
- Cheung, K.W.E.; Choi, S.R.; Lee, L.T.C.; Lee, N.L.E.; Tsang, H.F.; Cheng, Y.T.; Cho, W.C.S.; Wong, E.Y.L.; Wong, S.C.C. The potential of circulating cell free RNA as a biomarker in cancer. Expert Rev. Mol. Diagn. 2019, 19, 579–590. [Google Scholar] [CrossRef]
- Larson, M.H.; Pan, W.; Kim, H.J.; Mauntz, R.E.; Stuart, S.M.; Pimentel, M.; Zhou, Y.; Knudsgaard, P.; Demas, V.; Aravanis, A.M.; et al. A comprehensive characterization of the cell-free transcriptome reveals tissue- and subtype-specific biomarkers for cancer detection. Nat. Commun. 2021, 12, 2357. [Google Scholar] [CrossRef] [PubMed]
- Pezzicoli, G.; Tucci, M.; Lovero, D.; Silvestris, F.; Porta, C.; Mannavola, F. Large Extracellular Vesicles-A New Frontier of Liquid Biopsy in Oncology. Int. J. Mol. Sci. 2020, 21, 6543. [Google Scholar] [CrossRef]
- Weismann, P.; Weismanova, E.; Masak, L.; Mlada, K.; Keder, D.; Ferancikova, Z.; Vizvaryova, M.; Konecny, M.; Zavodna, K.; Kausitz, J.; et al. The detection of circulating tumor cells expressing E6/E7 HR-HPV oncogenes in peripheral blood in cervical cancer patients after radical hysterectomy. Neoplasma 2009, 56, 230–238. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Obermayr, E.; Sanchez-Cabo, F.; Tea, M.K.; Singer, C.F.; Krainer, M.; Fischer, M.B.; Sehouli, J.; Reinthaller, A.; Horvat, R.; Heinze, G.; et al. Assessment of a six gene panel for the molecular detection of circulating tumor cells in the blood of female cancer patients. BMC Cancer 2010, 10, 666. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kiss, I.; Kolostova, K.; Pawlak, I.; Bobek, V. Circulating tumor cells in gynaecological malignancies. J. BUON 2020, 25, 40–50. [Google Scholar]
- Du, K.; Huang, Q.; Bu, J.; Zhou, J.; Huang, Z.; Li, J. Circulating Tumor Cells Counting Act as a Potential Prognostic Factor in Cervical Cancer. Technol. Cancer Res. Treat. 2020, 19, 1533033820957005. [Google Scholar] [CrossRef]
- Stenman, J.; Lintula, S.; Hotakainen, K.; Vartiainen, J.; Lehvaslaiho, H.; Stenman, U.H. Detection of squamous-cell carcinoma antigen-expressing tumour cells in blood by reverse transcriptase-polymerase chain reaction in cancer of the uterine cervix. Int. J. Cancer 1997, 74, 75–80. [Google Scholar] [CrossRef]
- Pao, C.C.; Hor, J.J.; Yang, F.P.; Lin, C.Y.; Tseng, C.J. Detection of human papillomavirus mRNA and cervical cancer cells in peripheral blood of cervical cancer patients with metastasis. J. Clin. Oncol. 1997, 15, 1008–1012. [Google Scholar] [CrossRef] [Green Version]
- Yuan, C.C.; Wang, P.H.; Ng, H.T.; Li, Y.F.; Huang, T.S.; Chen, C.Y.; Tsai, L.C.; Shyong, W.Y. Detecting cytokeratin 19 mRNA in the peripheral blood cells of cervical cancer patients and its clinical-pathological correlation. Gynecol. Oncol. 2002, 85, 148–153. [Google Scholar] [CrossRef] [PubMed]
- Pfitzner, C.; Schroder, I.; Scheungraber, C.; Dogan, A.; Runnebaum, I.B.; Durst, M.; Hafner, N. Digital-Direct-RT-PCR: A sensitive and specific method for quantification of CTC in patients with cervical carcinoma. Sci. Rep. 2014, 4, 3970. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kolostova, K.; Spicka, J.; Matkowski, R.; Bobek, V. Isolation, primary culture, morphological and molecular characterization of circulating tumor cells in gynecological cancers. Am. J. Transl. Res. 2015, 7, 1203–1213. [Google Scholar]
- Takakura, M.; Matsumoto, T.; Nakamura, M.; Mizumoto, Y.; Myojyo, S.; Yamazaki, R.; Iwadare, J.; Bono, Y.; Orisaka, S.; Obata, T.; et al. Detection of circulating tumor cells in cervical cancer using a conditionally replicative adenovirus targeting telomerase-positive cells. Cancer Sci. 2018, 109, 231–240. [Google Scholar] [CrossRef] [Green Version]
- Wen, Y.F.; Cheng, T.T.; Chen, X.L.; Huang, W.J.; Peng, H.H.; Zhou, T.C.; Lin, X.D.; Zeng, L.S. Elevated circulating tumor cells and squamous cell carcinoma antigen levels predict poor survival for patients with locally advanced cervical cancer treated with radiotherapy. PLoS ONE 2018, 13, e0204334. [Google Scholar] [CrossRef]
- Pan, L.; Yan, G.; Chen, W.; Sun, L.; Wang, J.; Yang, J. Distribution of circulating tumor cell phenotype in early cervical cancer. Cancer Manag. Res. 2019, 11, 5531–5536. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tewari, K.S.; Sill, M.W.; Monk, B.J.; Penson, R.T.; Moore, D.H.; Lankes, H.A.; Ramondetta, L.M.; Landrum, L.M.; Randall, L.M.; Oaknin, A.; et al. Circulating Tumor Cells In Advanced Cervical Cancer: NRG Oncology-Gynecologic Oncology Group Study 240 (NCT 00803062). Mol. Cancer Ther. 2020, 19, 240. [Google Scholar] [CrossRef]
- Han, D.; Chen, K.; Che, J.; Hang, J.; Li, H. Detection of Epithelial-Mesenchymal Transition Status of Circulating Tumor Cells in Patients with Esophageal Squamous Carcinoma. Biomed. Res. Int. 2018, 2018, 7610154. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zeng, Y.E.; Yao, X.H.; Yan, Z.P.; Liu, J.X.; Liu, X.H. Potential signaling pathway involved in sphingosine-1-phosphate-induced epithelial-mesenchymal transition in cancer. Oncol. Lett. 2016, 12, 379–382. [Google Scholar] [CrossRef] [Green Version]
- Qi, L.N.; Xiang, B.D.; Wu, F.X.; Ye, J.Z.; Zhong, J.H.; Wang, Y.Y.; Chen, Y.Y.; Chen, Z.S.; Ma, L.; Chen, J.; et al. Circulating Tumor Cells Undergoing EMT Provide a Metric for Diagnosis and Prognosis of Patients with Hepatocellular Carcinoma. Cancer Res. 2018, 78, 4731–4744. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Yao, Y.; Xu, Y.; Li, L.; Gong, Y.; Zhang, K.; Zhang, M.; Guan, Y.; Chang, L.; Xia, X.; et al. Pan-cancer circulating tumor DNA detection in over 10,000 Chinese patients. Nat. Commun. 2021, 12, 11. [Google Scholar] [CrossRef]
- Cheung, T.H.; Yim, S.F.; Yu, M.Y.; Worley, M.J., Jr.; Fiascone, S.J.; Chiu, R.W.K.; Lo, K.W.K.; Siu, N.S.S.; Wong, M.C.S.; Yeung, A.C.M.; et al. Liquid biopsy of HPV DNA in cervical cancer. J. Clin. Virol. 2019, 114, 32–36. [Google Scholar] [CrossRef]
- Charo, L.M.; Eskander, R.N.; Okamura, R.; Patel, S.P.; Nikanjam, M.; Lanman, R.B.; Piccioni, D.E.; Kato, S.; McHale, M.T.; Kurzrock, R. Clinical implications of plasma circulating tumor DNA in gynecologic cancer patients. Mol. Oncol. 2021, 15, 67–79. [Google Scholar] [CrossRef] [PubMed]
- Liao, L.C.; Meng, Y.X.; Xian, A.L. Relationship between circulating tumor DNA in plasma and clinicopathology, effiacy and prognosis of cervical cancer. J. N. Sichuan Med. 2019, 34, 411–414. [Google Scholar]
- Kim, S.H.; Wu, M.; Stylianou, A.; Ghafoor, S.; Lakhman, Y.; Park, K.J., Jr.; Leitao, M.M.; Sonoda, Y.; Gardner, G.J.; Broach, V.; et al. Circulating cell-free DNA in patients with newly diagnosed and recurrent cervical cancer. Gynecol. Oncol. 2020, 159, 33–34. [Google Scholar] [CrossRef]
- Helen, F.G.C.C.; The Cancer Genome Atlas Research Network; Albert Einstein College of Medicine; Analytical Biological Services; Barretos Cancer Hospital; Baylor College of Medicine; Beckman Research Institute of City of Hope; Buck Institute for Research on Aging; Canada’s Michael Smith Genome Sciences Centre; Harvard Medical School; et al. Integrated genomic and molecular characterization of cervical cancer. Nature 2017, 543, 378–384. [Google Scholar] [CrossRef]
- Chung, T.K.H.; Cheung, T.H.; Yim, S.F.; Yu, M.Y.; Chiu, R.W.K.; Lo, K.W.K.; Lee, I.P.C.; Wong, R.R.Y.; Lau, K.K.M.; Wang, V.W.; et al. Liquid biopsy of PIK3CA mutations in cervical cancer in Hong Kong Chinese women. Gynecol. Oncol. 2017, 146, 334–339. [Google Scholar] [CrossRef]
- Tian, J.; Geng, Y.; Lv, D.; Li, P.; Cordova, M.; Liao, Y.; Tian, X.; Zhang, X.; Zhang, Q.; Zou, K.; et al. Using plasma cell-free DNA to monitor the chemoradiotherapy course of cervical cancer. Int. J. Cancer 2019, 145, 2547–2557. [Google Scholar] [CrossRef]
- Lee, S.Y.; Chae, D.K.; Lee, S.H.; Lim, Y.; An, J.; Chae, C.H.; Kim, B.C.; Bhak, J.; Bolser, D.; Cho, D.H. Efficient mutation screening for cervical cancers from circulating tumor DNA in blood. BMC Cancer 2020, 20, 694. [Google Scholar] [CrossRef]
- Tian, X.; Ge, D.; Zhang, F.; Zhang, B.; Bai, W.; Xu, X.; Li, Z.; Cao, Y.; Li, P.; Zou, K.; et al. Dynamic analysis of circulating tumor DNA to predict prognosis and monitor therapeutic response in metastatic relapsed cervical cancer. Int. J. Cancer 2021, 148, 921–931. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.Y.; Wei, S.J.; Lin, Y.C.; Lung, J.C.; Chang, T.C.; Whang-Peng, J.; Liu, J.M.; Yang, D.M.; Yang, W.K.; Shen, C.Y. PIK3CA as an oncogene in cervical cancer. Oncogene 2000, 19, 2739–2744. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, S.Y.; Chae, D.K.; An, J.; Yoo, S.; Jung, S.; Chae, C.H.; Bhak, J.; Kim, B.C.; Cho, D.H. Combinatory Analysis of Cell-free and Circulating Tumor Cell DNAs Provides More Variants for Cancer Treatment. Anticancer. Res. 2019, 39, 6595–6602. [Google Scholar] [CrossRef]
- Carow, K.; Golitz, M.; Wolf, M.; Hafner, N.; Jansen, L.; Hoyer, H.; Schwarz, E.; Runnebaum, I.B.; Durst, M. Viral-Cellular DNA Junctions as Molecular Markers for Assessing Intra-Tumor Heterogeneity in Cervical Cancer and for the Detection of Circulating Tumor DNA. Int. J. Mol. Sci. 2017, 18, 2032. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gu, Y.; Wan, C.; Qiu, J.; Cui, Y.; Jiang, T.; Zhuang, Z. Circulating HPV cDNA in the blood as a reliable biomarker for cervical cancer: A meta-analysis. PLoS ONE 2020, 15, e0224001. [Google Scholar] [CrossRef] [PubMed]
- Guerrero-Preston, R.; Valle, B.L.; Jedlicka, A.; Turaga, N.; Folawiyo, O.; Pirini, F.; Lawson, F.; Vergura, A.; Noordhuis, M.; Dziedzic, A.; et al. Molecular Triage of Premalignant Lesions in Liquid-Based Cervical Cytology and Circulating Cell-Free DNA from Urine, Using a Panel of Methylated Human Papilloma Virus and Host Genes. Cancer Prev. Res. 2016, 9, 915–924. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Berti, F.C.B.; Salviano-Silva, A.; Beckert, H.C.; de Oliveira, K.B.; Cipolla, G.A.; Malheiros, D. From squamous intraepithelial lesions to cervical cancer: Circulating microRNAs as potential biomarkers in cervical carcinogenesis. Biochim. Biophys. Acta Rev. Cancer 2019, 1872, 188306. [Google Scholar] [CrossRef] [PubMed]
- Jansen, R.P. mRNA localization: Message on the move. Nat. Rev. Mol. Cell. Biol. 2001, 2, 247–256. [Google Scholar] [CrossRef]
- Tseng, C.J.; Pao, C.C.; Lin, J.D.; Soong, Y.K.; Hong, J.H.; Hsueh, S. Detection of human papillomavirus types 16 and 18 mRNA in peripheral blood of advanced cervical cancer patients and its association with prognosis. J. Clin. Oncol. 1999, 17, 1391–1396. [Google Scholar] [CrossRef]
- Mitsuhashi, A.; Tanaka, N.; Suzuka, K.; Matsui, H.; Seki, K.; Sekiya, S. Detection of epidermal growth factor receptor mRNA in peripheral blood of cervical cancer patients. Gynecol. Oncol. 2003, 89, 480–485. [Google Scholar] [CrossRef]
- Zhang, X.; Wang, C.; Wang, L.; Du, L.; Wang, S.; Zheng, G.; Li, W.; Zhuang, X.; Zhang, X.; Dong, Z. Detection of circulating Bmi-1 mRNA in plasma and its potential diagnostic and prognostic value for uterine cervical cancer. Int. J. Cancer 2012, 131, 165–172. [Google Scholar] [CrossRef]
- Reddi, K.K.; Holland, J.F. Elevated serum ribonuclease in patients with pancreatic cancer. Proc. Natl. Acad. Sci. USA 1976, 73, 2308–2310. [Google Scholar] [CrossRef] [Green Version]
- Rapisuwon, S.; Vietsch, E.E.; Wellstein, A. Circulating biomarkers to monitor cancer progression and treatment. Comput. Struct. Biotechnol. J. 2016, 14, 211–222. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Esteller, M. Non-coding RNAs in human disease. Nat. Rev. Genet. 2011, 12, 861–874. [Google Scholar] [CrossRef] [PubMed]
- Tornesello, M.L.; Faraonio, R.; Buonaguro, L.; Annunziata, C.; Starita, N.; Cerasuolo, A.; Pezzuto, F.; Tornesello, A.L.; Buonaguro, F.M. The Role of microRNAs, Long Non-coding RNAs, and Circular RNAs in Cervical Cancer. Front. Oncol. 2020, 10, 150. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, W.; Wang, L.; Zhao, D.; Wang, P.; Li, Y.; Wang, S. Four Circulating Long Non-Coding RNAs Act as Biomarkers for Predicting Cervical Cancer. Gynecol. Obstet. Invest. 2018, 83, 533–539. [Google Scholar] [CrossRef]
- Iempridee, T.; Wiwithaphon, S.; Piboonprai, K.; Pratedrat, P.; Khumkhrong, P.; Japrung, D.; Temisak, S.; Laiwejpithaya, S.; Chaopotong, P.; Dharakul, T. Identification of reference genes for circulating long noncoding RNA analysis in serum of cervical cancer patients. FEBS Open Bio 2018, 8, 1844–1854. [Google Scholar] [CrossRef]
- Ding, X.Z.; Zhang, S.Q.; Deng, X.L.; Qiang, J.H. Serum Exosomal lncRNA DLX6-AS1 Is a Promising Biomarker for Prognosis Prediction of Cervical Cancer. Technol. Cancer Res. Treat. 2021, 20, 1533033821990060. [Google Scholar] [CrossRef]
- Yu, J.; Wang, Y.; Dong, R.; Huang, X.; Ding, S.; Qiu, H. Circulating microRNA-218 was reduced in cervical cancer and correlated with tumor invasion. J. Cancer Res. Clin. Oncol. 2012, 138, 671–674. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.; Yao, D.; Chen, J.; Ding, N. Circulating miRNA-20a and miRNA-203 for screening lymph node metastasis in early stage cervical cancer. Genet. Test. Mol. Biomarkers 2013, 17, 631–636. [Google Scholar] [CrossRef]
- Chen, J.; Yao, D.; Li, Y.; Chen, H.; He, C.; Ding, N.; Lu, Y.; Ou, T.; Zhao, S.; Li, L.; et al. Serum microRNA expression levels can predict lymph node metastasis in patients with early-stage cervical squamous cell carcinoma. Int. J. Mol. Med. 2013, 32, 557–567. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, P.; Xin, F.; Ma, C.F. Clinical significance of serum miR-196a in cervical intraepithelial neoplasia and cervical cancer. Genet. Mol. Res. 2015, 14, 17995–18002. [Google Scholar] [CrossRef]
- Jia, W.; Wu, Y.; Zhang, Q.; Gao, G.E.; Zhang, C.; Xiang, Y. Expression profile of circulating microRNAs as a promising fingerprint for cervical cancer diagnosis and monitoring. Mol. Clin. Oncol. 2015, 3, 851–858. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, N.; Fei, D.; Zong, S.; Zhang, M.; Yue, Y. MicroRNA-138 inhibits proliferation, migration and invasion through targeting hTERT in cervical cancer. Oncol. Lett. 2016, 12, 3633–3639. [Google Scholar] [CrossRef] [PubMed]
- Mou, Z.; Xu, X.; Dong, M.; Xu, J. MicroRNA-148b Acts as a Tumor Suppressor in Cervical Cancer by Inducing G1/S-Phase Cell Cycle Arrest and Apoptosis in a Caspase-3-Dependent Manner. Med. Sci. Monit. 2016, 22, 2809–2815. [Google Scholar] [CrossRef] [Green Version]
- Sun, L.; Jiang, R.; Li, J.; Wang, B.; Ma, C.; Lv, Y.; Mu, N. MicoRNA-425-5p is a potential prognostic biomarker for cervical cancer. Ann. Clin. Biochem. 2017, 54, 127–133. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, H.; Chen, J.; Li, D.; Liu, X.; Li, L.; Wang, K. MicroRNA-30e Functions as a Tumor Suppressor in Cervical Carcinoma Cells through Targeting GALNT7. Transl. Oncol. 2017, 10, 876–885. [Google Scholar] [CrossRef]
- Liang, H.; Luo, R.; Chen, X.; Zhao, Y.; Tan, A. miR-187 inhibits the growth of cervical cancer cells by targeting FGF9. Oncol. Rep. 2017, 38, 1977–1984. [Google Scholar] [CrossRef] [Green Version]
- Li, H.; Sheng, Y.; Zhang, Y.; Gao, N.; Deng, X.; Sheng, X. MicroRNA-138 is a potential biomarker and tumor suppressor in human cervical carcinoma by reversely correlated with TCF3 gene. Gynecol. Oncol. 2017, 145, 569–576. [Google Scholar] [CrossRef] [PubMed]
- Song, R.; Cong, L.; Ni, G.; Chen, M.; Sun, H.; Sun, Y.; Chen, M. MicroRNA-195 inhibits the behavior of cervical cancer tumors by directly targeting HDGF. Oncol. Lett. 2017, 14, 767–775. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.M.; Ju, B.H.; Pan, C.J.; Gu, Y.; Li, M.Q.; Sun, L.; Xu, Y.Y.; Yin, L.R. MiR-214 inhibits cell migration, invasion and promotes the drug sensitivity in human cervical cancer by targeting FOXM1. Am. J. Transl. Res. 2017, 9, 3541–3557. [Google Scholar] [PubMed]
- Li, C.; Zheng, X.; Li, W.; Bai, F.; Lyu, J.; Meng, Q.H. Serum miR-486-5p as a diagnostic marker in cervical cancer: With investigation of potential mechanisms. BMC Cancer 2018, 18, 61. [Google Scholar] [CrossRef] [Green Version]
- Shukla, V.; Varghese, V.K.; Kabekkodu, S.P.; Mallya, S.; Chakrabarty, S.; Jayaram, P.; Pandey, D.; Banerjee, S.; Sharan, K.; Satyamoorthy, K. Enumeration of deregulated miRNAs in liquid and tissue biopsies of cervical cancer. Gynecol. Oncol. 2019, 155, 135–143. [Google Scholar] [CrossRef]
- Du, S.; Zhao, Y.; Lv, C.; Wei, M.; Gao, Z.; Meng, X. Applying Serum Proteins and MicroRNA as Novel Biomarkers for Early-Stage Cervical Cancer Detection. Sci. Rep. 2020, 10, 9033. [Google Scholar] [CrossRef] [PubMed]
- Zheng, M.; Hou, L.; Ma, Y.; Zhou, L.; Wang, F.; Cheng, B.; Wang, W.; Lu, B.; Liu, P.; Lu, W.; et al. Exosomal let-7d-3p and miR-30d-5p as diagnostic biomarkers for non-invasive screening of cervical cancer and its precursors. Mol. Cancer 2019, 18, 76. [Google Scholar] [CrossRef] [PubMed]
- Lv, A.; Tu, Z.; Huang, Y.; Lu, W.; Xie, B. Circulating exosomal miR-125a-5p as a novel biomarker for cervical cancer. Oncol. Lett. 2021, 21, 54. [Google Scholar] [CrossRef] [PubMed]
- Mannavola, F.; D’Oronzo, S.; Cives, M.; Stucci, L.S.; Ranieri, G.; Silvestris, F.; Tucci, M. Extracellular Vesicles and Epigenetic Modifications Are Hallmarks of Melanoma Progression. Int. J. Mol. Sci. 2019, 21, 52. [Google Scholar] [CrossRef] [Green Version]
- Barwal, T.S.; Sharma, U.; Vasquez, K.M.; Prakash, H.; Jain, A. A panel of circulating long non-coding RNAs as liquid biopsy biomarkers for breast and cervical cancers. Biochimie 2020, 176, 62–70. [Google Scholar] [CrossRef]
- Tang, Q.; Hann, S.S. HOTAIR: An Oncogenic Long Non-Coding RNA in Human Cancer. Cell. Physiol. Biochem. 2018, 47, 893–913. [Google Scholar] [CrossRef]
- Cai, B.; Song, X.Q.; Cai, J.P.; Zhang, S. HOTAIR: A cancer-related long non-coding RNA. Neoplasma 2014, 61, 379–391. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Cheng, X.; Liang, H.; Jin, Z. Long non-coding RNA HOTAIR and STAT3 synergistically regulate the cervical cancer cell migration and invasion. Chem. Biol. Interact. 2018, 286, 106–110. [Google Scholar] [CrossRef]
- Guo, J.; Chen, Z.; Jiang, H.; Yu, Z.; Peng, J.; Xie, J.; Li, Z.; Wu, W.; Cheng, Z.; Xiao, K. The lncRNA DLX6-AS1 promoted cell proliferation, invasion, migration and epithelial-to-mesenchymal transition in bladder cancer via modulating Wnt/beta-catenin signaling pathway. Cancer Cell Int. 2019, 19, 312. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, P.; Guan, H.; Dai, Z.; Ma, Y.; Zhao, Y.; Liu, D. Long noncoding RNA DLX6-AS1 promotes breast cancer progression via miR-505-3p/RUNX2 axis. Eur. J. Pharmacol. 2019, 865, 172778. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Li, Y.; Yang, Y.; Liu, X.; Zang, M.; Li, Y.; Yang, K.; Yang, W.; Zhang, S. Long noncoding RNA DLX6AS1 is associated with malignant progression and promotes proliferation and invasion in esophageal squamous cell carcinoma. Mol. Med. Rep. 2019, 19, 1942–1950. [Google Scholar] [CrossRef] [Green Version]
- Sun, W.; Zhang, L.; Yan, R.; Yang, Y.; Meng, X. LncRNA DLX6-AS1 promotes the proliferation, invasion, and migration of non-small cell lung cancer cells by targeting the miR-27b-3p/GSPT1 axis. OncoTargets Ther. 2019, 12, 3945–3954. [Google Scholar] [CrossRef] [Green Version]
- Peng, Y.; Croce, C.M. The role of MicroRNAs in human cancer. Signal Transduct. Target. Ther. 2016, 1, 15004. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mori, M.A.; Ludwig, R.G.; Garcia-Martin, R.; Brandao, B.B.; Kahn, C.R. Extracellular miRNAs: From Biomarkers to Mediators of Physiology and Disease. Cell Metab. 2019, 30, 656–673. [Google Scholar] [CrossRef]
- He, Y.; Lin, J.; Ding, Y.; Liu, G.; Luo, Y.; Huang, M.; Xu, C.; Kim, T.K.; Etheridge, A.; Lin, M.; et al. A systematic study on dysregulated microRNAs in cervical cancer development. Int. J. Cancer 2016, 138, 1312–1327. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Uesugi, A.; Kozaki, K.; Tsuruta, T.; Furuta, M.; Morita, K.; Imoto, I.; Omura, K.; Inazawa, J. The tumor suppressive microRNA miR-218 targets the mTOR component Rictor and inhibits AKT phosphorylation in oral cancer. Cancer Res. 2011, 71, 5765–5778. [Google Scholar] [CrossRef] [Green Version]
- Tie, J.; Pan, Y.; Zhao, L.; Wu, K.; Liu, J.; Sun, S.; Guo, X.; Wang, B.; Gang, Y.; Zhang, Y.; et al. MiR-218 inhibits invasion and metastasis of gastric cancer by targeting the Robo1 receptor. PLoS Genet. 2010, 6, e1000879. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Zheng, F.; Yu, G.; Yin, Y.; Lu, Q. miR-196a targets netrin 4 and regulates cell proliferation and migration of cervical cancer cells. Biochem. Biophys. Res. Commun. 2013, 440, 582–588. [Google Scholar] [CrossRef] [PubMed]
- Hou, T.; Ou, J.; Zhao, X.; Huang, X.; Huang, Y.; Zhang, Y. MicroRNA-196a promotes cervical cancer proliferation through the regulation of FOXO1 and p27Kip1. Br. J. Cancer 2014, 110, 1260–1268. [Google Scholar] [CrossRef] [Green Version]
- Villegas-Ruiz, V.; Juarez-Mendez, S.; Perez-Gonzalez, O.A.; Arreola, H.; Paniagua-Garcia, L.; Parra-Melquiadez, M.; Peralta-Rodriguez, R.; Lopez-Romero, R.; Monroy-Garcia, A.; Mantilla-Morales, A.; et al. Heterogeneity of microRNAs expression in cervical cancer cells: Over-expression of miR-196a. Int. J. Clin. Exp. Pathol. 2014, 7, 1389–1401. [Google Scholar]
- Sharma, S.; Mandal, P.; Sadhukhan, T.; Roy Chowdhury, R.; Ranjan Mondal, N.; Chakravarty, B.; Chatterjee, T.; Roy, S.; Sengupta, S. Bridging Links between Long Noncoding RNA HOTAIR and HPV Oncoprotein E7 in Cervical Cancer Pathogenesis. Sci. Rep. 2015, 5, 11724. [Google Scholar] [CrossRef] [Green Version]
- Condrat, C.E.; Thompson, D.C.; Barbu, M.G.; Bugnar, O.L.; Boboc, A.; Cretoiu, D.; Suciu, N.; Cretoiu, S.M.; Voinea, S.C. miRNAs as Biomarkers in Disease: Latest Findings Regarding Their Role in Diagnosis and Prognosis. Cells 2020, 9, 276. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, H.; Peng, R.; Wang, J.; Qin, Z.; Xue, L. Circulating microRNAs as potential cancer biomarkers: The advantage and disadvantage. Clin. Epigenetics 2018, 10, 59. [Google Scholar] [CrossRef] [Green Version]
- Hasanzadeh, M.; Movahedi, M.; Rejali, M.; Maleki, F.; Moetamani-Ahmadi, M.; Seifi, S.; Hosseini, Z.; Khazaei, M.; Amerizadeh, F.; Ferns, G.A.; et al. The potential prognostic and therapeutic application of tissue and circulating microRNAs in cervical cancer. J. Cell. Physiol. 2019, 234, 1289–1294. [Google Scholar] [CrossRef]
- Rashed, M.H.; Bayraktar, E.; Helal, G.K.; Abd-Ellah, M.F.; Amero, P.; Chavez-Reyes, A.; Rodriguez-Aguayo, C. Exosomes: From Garbage Bins to Promising Therapeutic Targets. Int. J. Mol. Sci. 2017, 18, 538. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iorio, M.V.; Ferracin, M.; Liu, C.G.; Veronese, A.; Spizzo, R.; Sabbioni, S.; Magri, E.; Pedriali, M.; Fabbri, M.; Campiglio, M.; et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005, 65, 7065–7070. [Google Scholar] [CrossRef] [Green Version]
- Jiang, L.; Huang, Q.; Chang, J.; Wang, E.; Qiu, X. MicroRNA HSA-miR-125a-5p induces apoptosis by activating p53 in lung cancer cells. Exp. Lung Res. 2011, 37, 387–398. [Google Scholar] [CrossRef]
| Method | Markers for CTC Detection | N. of Patients | Clinical Significance | Reference |
|---|---|---|---|---|
| Reverse transcriptase-polymerase chain reaction (RT-PCR) | Squamous-cell carcinoma (SCC) antigen mRNA | 15 | Prognostic | [30] |
| RT-PCR | HPV type 16 E6 mRNA | 15 | Prognostic | [31] |
| Nested RT-PCR | CK19 mRNA | 84 | None | [32] |
| Digital-direct-RT-PCR | HPV16, HPV18 mRNA | 10 | None | [33] |
| Filtration through 8μm membrane pores and in vitro culture of CTCs | Cytomorphological evaluation and gene expression profiling | 1 | None | [34] |
| Peripheral blood cell infection with a green fluorescent protein (GFP)-modified adenoviral vector (OBP-1101) and subsequent fluorescence imaging and capture of GFP+/CD45− CTC | E6/E7 HPV gene | 23 | None | [35] |
| Negative enrichment with anti-CD45 microbeads and fluorescence in situ hybridization (FISH) for CEP8 probe | Hyperdiploid CEP8+/DAPI+/CD45- | 99 | Prognostic | [36] |
| CanPatrolTM technique and anti-CD45 antibody | RNA in situ hybridization for epithelial (EPCAM, CK8) and mesenchymal (Vimentin, TWIST) markers | 90 | Prognostic | [37] |
| Negative enrichment with anti-CD45 microbeads and FISH for CEP8 probe | Hyperdiploid CEP8+/DAPI+/CD45− | 107 | Prognostic | [29] |
| CellSearch system | Pan-CK+/CD45− | 176 | Predictive | [38] |
| Mutated Genes/Viral DNA | Method | N. of CC Patients | Putative Clinical Validity | References |
|---|---|---|---|---|
| PIK3CA | dPCR | 117 | Prognostic | [48] |
| ALK, RET, CSF1R, MET, EGFR, APC, ABL1, NOTCH1, KDR, HNF1A, PDGFRA, ATM, SMO, ERBB2, FGFR2, GNAS, TP53, PTPN11, KRAS, CDH1, FLT3, FGFR3, MLH1, PIK3CA, PTEN, JAK3, MPL, ERBB4, KIT, RB1, IDH1 | NGS | 57 | Prognostic | [49] |
| ZFHX3, KMT2C, KMT2D, NSD1, ATM, RNF213, FAT1, CHD4, FAT4, TRRAP, EP300, PIK3CA, PTEN, TP53, ARID1A, CTCF, PIK3R1, FXBW7 | NGS | 24 | Follow-up | [50] |
| PIK3CA, TP53, FXBW7, ERBB2, PTEN, CDKN2A, KRAS, BRAF, MYC, MET, ARID1A, CCNE1, FCFR2, APC, CTNNB1, NRAS, CCND1, TERT | NGS | 13 | Prognostic, Predictive | [44] |
| PIK3CA, BRAF, GNA11, FBXW7, CDH1, ALK, STK11, VHL, PDGFRA, HNF1A, MPL, ABL1, RET, KDR, KIT, CDFR1, ATM, EGFR, FGFR1, FGFR2, GNAS, AKT1, KRAS, PTEN, SRC, FLT3, SMO, HRAS, JAK3 | NGS | 82 | Prognostic, Follow-up | [51] |
| PIK3CA (30.1%), MLL3, TP53, MLL2, EP300, PTEN, FGFR3, DNMT3A, PTCH1, TERT, AKT1, BRAF, BRCA1, ERBB2, TSC2 | NGS | 126 | Prognostic, predictive | [42] |
| Method | Advantages | Disadvantages | Clinical Applications |
|---|---|---|---|
| Circulating tumor cells (CTCs) |
|
|
|
| Circulating tumor DNA (ctDNA) |
|
|
|
| Circulating Coding RNAs | • Early detection of cancer and disease monitoring |
| |
| Circulating Non-coding RNAs |
|
| |
| Exosomal miRNAs |
|
| • Diagnostic biomarkers [108,109,110] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cafforio, P.; Palmirotta, R.; Lovero, D.; Cicinelli, E.; Cormio, G.; Silvestris, E.; Porta, C.; D’Oronzo, S. Liquid Biopsy in Cervical Cancer: Hopes and Pitfalls. Cancers 2021, 13, 3968. https://doi.org/10.3390/cancers13163968
Cafforio P, Palmirotta R, Lovero D, Cicinelli E, Cormio G, Silvestris E, Porta C, D’Oronzo S. Liquid Biopsy in Cervical Cancer: Hopes and Pitfalls. Cancers. 2021; 13(16):3968. https://doi.org/10.3390/cancers13163968
Chicago/Turabian StyleCafforio, Paola, Raffaele Palmirotta, Domenica Lovero, Ettore Cicinelli, Gennaro Cormio, Erica Silvestris, Camillo Porta, and Stella D’Oronzo. 2021. "Liquid Biopsy in Cervical Cancer: Hopes and Pitfalls" Cancers 13, no. 16: 3968. https://doi.org/10.3390/cancers13163968
APA StyleCafforio, P., Palmirotta, R., Lovero, D., Cicinelli, E., Cormio, G., Silvestris, E., Porta, C., & D’Oronzo, S. (2021). Liquid Biopsy in Cervical Cancer: Hopes and Pitfalls. Cancers, 13(16), 3968. https://doi.org/10.3390/cancers13163968