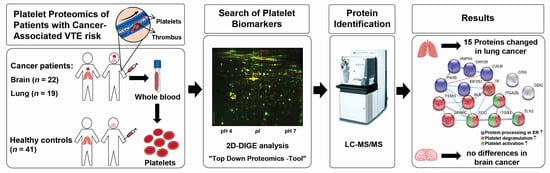
Alterations of the Platelet Proteome in Lung Cancer: Accelerated F13A1 and ER Processing as New Actors in Hypercoagulability
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Design
2.2. Blood Collection, Washed Platelet and Plasma Isolation
2.3. Platelet Preparation for Two-Dimensional Fluorescence Differential Gel Electrophoresis (2D-DIGE) Analysis
2.4. Platelet Proteome Analysis by 2D-DIGE
2.5. 2D-DIGE Image Analysis
2.6. Protein Identification via Mass Spectrometry
2.7. One- and Two-Dimensional Western Blot Analysis
2.8. Measurement of Haemostatic Biomarkers in Plasma
2.9. Fluorogenic F13A1 Activity Assay from Platelet Samples
2.10. Plasmin–α-2-Antiplasmin (PAP) Complex Quantification in Plasma by ELISA
2.11. Protein Disulfide Isomerase (P4HB) Quantification in Plasma by ELISA
2.12. Biological Pathway Analysis
2.13. Statistics
2.14. Control Evaluation of the Platelet Proteome Analysis in Comparison with the Different Platelet Counts between Study Groups
3. Results
3.1. Patient Characteristics
3.2. Platelet Proteome of Patients with Brain and Lung Cancer Compared to Controls
3.3. Pathway Analysis of Lung Cancer-Related Platelet Proteins
3.4. F13A1 Protein Processing Is Changed in Platelets of Patients with Lung Cancer
3.5. Chaperones from the KEGG Pathway “Protein Processing in ER” Are Elevated in Platelets of Patients with Lung Cancer
3.6. Functional Relationships of the Platelet Proteome with Haemostatic Plasma Laboratory Parameters from Patients with Lung Cancer and Matched Controls
3.7. Associations in the Platelet Proteome with Lung Cancer and Risk of Death
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Mulder, F.I.; Horvath-Puho, E.; van Es, N.; van Laarhoven, H.; Pedersen, L.; Moik, F.; Ay, C.; Buller, H.R.; Toft Sorensen, H. Venous thromboembolism in cancer patients: A population-based cohort study. Blood 2020, 137, 1959–1969. [Google Scholar] [CrossRef] [PubMed]
- Horsted, F.; West, J.; Grainge, M.J. Risk of venous thromboembolism in patients with cancer: A systematic review and meta-analysis. PLoS Med. 2012, 9, e1001275. [Google Scholar] [CrossRef]
- Ay, C.; Dunkler, D.; Pirker, R.; Thaler, J.; Quehenberger, P.; Wagner, O.; Zielinski, C.; Pabinger, I. High D-dimer levels are associated with poor prognosis in cancer patients. Haematologica 2012, 97, 1158–1164. [Google Scholar] [CrossRef] [Green Version]
- Moik, F.; Ay, C.; Pabinger, I. Risk prediction for cancer-associated thrombosis in ambulatory patients with cancer: Past, present and future. Thromb. Res. 2020, 191 (Suppl. 1), S3–S11. [Google Scholar] [CrossRef]
- Riedl, J.; Kaider, A.; Marosi, C.; Prager, G.W.; Eichelberger, B.; Assinger, A.; Pabinger, I.; Panzer, S.; Ay, C. Decreased platelet reactivity in patients with cancer is associated with high risk of venous thromboembolism and poor prognosis. Thromb. Haemost. 2017, 117, 90–98. [Google Scholar] [CrossRef]
- Grilz, E.; Konigsbrugge, O.; Posch, F.; Schmidinger, M.; Pirker, R.; Lang, I.M.; Pabinger, I.; Ay, C. Frequency, risk factors, and impact on mortality of arterial thromboembolism in patients with cancer. Haematologica 2018, 103, 1549–1556. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Simanek, R.; Vormittag, R.; Ay, C.; Alguel, G.; Dunkler, D.; Schwarzinger, I.; Steger, G.; Jaeger, U.; Zielinski, C.; Pabinger, I. High platelet count associated with venous thromboembolism in cancer patients: Results from the Vienna Cancer and Thrombosis Study (CATS). J. Thromb. Haemost. 2010, 8, 114–120. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Bi, Y.; Kou, J.; Shi, J.; Piao, D. Phosphatidylserine exposing-platelets and microparticles promote procoagulant activity in colon cancer patients. J. Exp. Clin. Cancer Res. 2016, 35, 54. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ay, C.; Jungbauer, L.V.; Sailer, T.; Tengler, T.; Koder, S.; Kaider, A.; Panzer, S.; Quehenberger, P.; Pabinger, I.; Mannhalter, C. High concentrations of soluble P-selectin are associated with risk of venous thromboembolism and the P-selectin Thr715 variant. Clin. Chem. 2007, 53, 1235–1243. [Google Scholar] [CrossRef] [Green Version]
- Nash, G.F.; Turner, L.F.; Scully, M.F.; Kakkar, A.K. Platelets and cancer. Lancet Oncol. 2002, 3, 425–430. [Google Scholar] [CrossRef]
- Camerer, E.; Qazi, A.A.; Duong, D.N.; Cornelissen, I.; Advincula, R.; Coughlin, S.R. Platelets, protease-activated receptors, and fibrinogen in hematogenous metastasis. Blood 2004, 104, 397–401. [Google Scholar] [CrossRef] [PubMed]
- Rothwell, P.M.; Fowkes, F.G.; Belch, J.F.; Ogawa, H.; Warlow, C.P.; Meade, T.W. Effect of daily aspirin on long-term risk of death due to cancer: Analysis of individual patient data from randomised trials. Lancet 2011, 377, 31–41. [Google Scholar] [CrossRef]
- Rothwell, P.M.; Wilson, M.; Price, J.F.; Belch, J.F.; Meade, T.W.; Mehta, Z. Effect of daily aspirin on risk of cancer metastasis: A study of incident cancers during randomised controlled trials. Lancet 2012, 379, 1591–1601. [Google Scholar] [CrossRef]
- Contursi, A.; Sacco, A.; Grande, R.; Dovizio, M.; Patrignani, P. Platelets as crucial partners for tumor metastasis: From mechanistic aspects to pharmacological targeting. Cell. Mol. Life Sci. 2017, 74, 3491–3507. [Google Scholar] [CrossRef] [PubMed]
- Strohkamp, S.; Gemoll, T.; Humborg, S.; Hartwig, S.; Lehr, S.; Freitag-Wolf, S.; Becker, S.; Franzen, B.; Pries, R.; Wollenberg, B.; et al. Protein levels of clusterin and glutathione synthetase in platelets allow for early detection of colorectal cancer. Cell. Mol. Life Sci. 2018, 75, 323–334. [Google Scholar] [CrossRef] [PubMed]
- Hell, L.; Lurger, K.; Mauracher, L.M.; Grilz, E.; Reumiller, C.M.; Schmidt, G.J.; Ercan, H.; Koder, S.; Assinger, A.; Basilio, J.; et al. Altered platelet proteome in lupus anticoagulant (LA)-positive patients-protein disulfide isomerase and NETosis as new players in LA-related thrombosis. Exp. Mol. Med. 2020, 52, 66–78. [Google Scholar] [CrossRef]
- Lu, X.; Wang, Z.; Gao, Y.; Chen, W.; Wang, L.; Huang, P.; Gao, W.; Ke, M.; He, A.; Tian, R. AutoProteome chip system for fully automated and integrated proteomics sample preparation and peptide fractionation. Anal. Chem. 2020, 92, 8893–8900. [Google Scholar] [CrossRef] [PubMed]
- Alexovic, M.; Urban, P.L.; Tabani, H.; Sabo, J. Recent advances in robotic protein sample preparation for clinical analysis and other biomedical applications. Clin. Chim. Acta Int. J. Clin. Chem. 2020, 507, 104–116. [Google Scholar] [CrossRef] [PubMed]
- Meleady, P. Two-Dimensional gel electrophoresis and 2D-DIGE. Methods Mol. Biol. 2018, 1664, 3–14. [Google Scholar] [CrossRef]
- Smith, L.M.; Kelleher, N.L.; Consortium for Top down Proteomics. Proteoform: A single term describing protein complexity. Nat. Methods 2013, 10, 186–187. [Google Scholar] [CrossRef] [Green Version]
- Tannu, N.S.; Hemby, S.E. Two-Dimensional fluorescence difference gel electrophoresis for comparative proteomics profiling. Nat. Protoc. 2006, 1, 1732–1742. [Google Scholar] [CrossRef] [PubMed]
- Riedl, J.; Hell, L.; Kaider, A.; Koder, S.; Marosi, C.; Zielinski, C.; Panzer, S.; Pabinger, I.; Ay, C. Association of platelet activation markers with cancer-associated venous thromboembolism. Platelets 2016, 27, 80–85. [Google Scholar] [CrossRef] [PubMed]
- Ay, C.; Simanek, R.; Vormittag, R.; Dunkler, D.; Alguel, G.; Koder, S.; Kornek, G.; Marosi, C.; Wagner, O.; Zielinski, C.; et al. High plasma levels of soluble P-selectin are predictive of venous thromboembolism in cancer patients: Results from the Vienna Cancer and Thrombosis Study (CATS). Blood 2008, 112, 2703–2708. [Google Scholar] [CrossRef] [Green Version]
- Alban, A.; David, S.O.; Bjorkesten, L.; Andersson, C.; Sloge, E.; Lewis, S.; Currie, I. A novel experimental design for comparative two-dimensional gel analysis: Two-dimensional difference gel electrophoresis incorporating a pooled internal standard. Proteomics 2003, 3, 36–44. [Google Scholar] [CrossRef]
- Winkler, W.; Zellner, M.; Diestinger, M.; Babeluk, R.; Marchetti, M.; Goll, A.; Zehetmayer, S.; Bauer, P.; Rappold, E.; Miller, I.; et al. Biological variation of the platelet proteome in the elderly population and its implication for biomarker research. Mol. Cell. Proteom. 2008, 7, 193–203. [Google Scholar] [CrossRef] [Green Version]
- Shevchenko, A.; Wilm, M.; Vorm, O.; Mann, M. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem. 1996, 68, 850–858. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, G.J.; Reumiller, C.M.; Ercan, H.; Resch, U.; Butt, E.; Heber, S.; Liutkeviciute, Z.; Basilio, J.; Schmid, J.A.; Assinger, A.; et al. Comparative proteomics reveals unexpected quantitative phosphorylation differences linked to platelet activation state. Sci. Rep. 2019, 9, 19009. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Szklarczyk, D.; Simonovic, M.; von Mering, C.; Forslund, K.; Bork, P.; Kuhn, M.; Lyon, D.; Cook, H.V.; Delgado, A.S.; Jensen, E.J. STRING is a Database of Known and Predicted Protein-Protein Interactions. Available online: https://string-db.org/cgi/about?footer_active_subpage=contributors (accessed on 26 October 2020).
- Zhou, G.; Soufan, O.; Ewald, J.; Hancock, R.E.W.; Basu, N.; Xia, J. NetworkAnalyst 3.0: A visual analytics platform for comprehensive gene expression profiling and meta-analysis. Nucleic Acids Res. 2019, 47, W234–W241. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, G.; Soufan, O.; Ewald, J.; Hancock, R.E.W.; Basu, N.; Xia, J. NetworkAnalyst—A Comprehensive Network Visual Analytics Platform for Gene Expression Analysis. Available online: https://www.networkanalyst.ca/ (accessed on 14 January 2021).
- Breuer, K.; Foroushani, A.K.; Laird, M.R.; Chen, C.; Sribnaia, A.; Lo, R.; Winsor, G.L.; Hancock, R.E.; Brinkman, F.S.; Lynn, D.J. InnateDB: Systems biology of innate immunity and beyond—Recent updates and continuing curation. Nucleic Acids Res. 2013, 41, D1228–D1233. [Google Scholar] [CrossRef]
- Benjamin, Y.; Hochberg, Y. On the adaptive control of the false discovery rate in multiple testing with independent statistics. J. Educ. Behav. Stat. 2000, 25, 60–83. [Google Scholar] [CrossRef] [Green Version]
- Radua, J.; Fusar-Poli, P.; Mataix-Cols, D.; Albajes-Eizagirre, A.; Rubie, K.; Thiebaut de Schotten, M. Online Calculator of FDR Correction for Multiple Comparisons. Available online: https://www.sdmproject.com/utilities/?show=FDR (accessed on 20 March 2020).
- Pipe, S.W.; Morris, J.A.; Shah, J.; Kaufman, R.J. Differential interaction of coagulation factor VIII and factor V with protein chaperones calnexin and calreticulin. J. Biol. Chem. 1998, 273, 8537–8544. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaufman, R.J.; Pipe, S.W.; Tagliavacca, L.; Swaroop, M.; Moussalli, M. Biosynthesis, assembly and secretion of coagulation factor VIII. Blood Coagul. Fibrinolysis 1997, 8 (Suppl. 2), S3–S14. [Google Scholar] [PubMed]
- Lorand, L.; Konishi, K. Activation of the fibrin stabilizing factor of plasma by thrombin. Arch. Biochem. Biophys. 1964, 105, 58–67. [Google Scholar] [CrossRef]
- Takagi, T.; Doolittle, R.F. Amino acid sequence studies on factor XIII and the peptide released during its activation by thrombin. Biochemistry 1974, 13, 750–756. [Google Scholar] [CrossRef]
- Takahashi, N.; Takahashi, Y.; Putnam, F.W. Primary structure of blood coagulation factor XIIIa (fibrinoligase, transglutaminase) from human placenta. Proc. Natl. Acad. Sci. USA 1986, 83, 8019–8023. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shubin, N.J.; Glukhova, V.A.; Clauson, M.; Truong, P.; Abrink, M.; Pejler, G.; White, N.J.; Deutsch, G.H.; Reeves, S.R.; Vaisar, T.; et al. Proteome analysis of mast cell releasates reveals a role for chymase in the regulation of coagulation factor XIIIA levels via proteolytic degradation. J. Allergy Clin. Immunol. 2017, 139, 323–334. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hur, W.S.; Mazinani, N.; Lu, X.J.; Britton, H.M.; Byrnes, J.R.; Wolberg, A.S.; Kastrup, C.J. Coagulation factor XIIIa is inactivated by plasmin. Blood 2015, 126, 2329–2337. [Google Scholar] [CrossRef] [Green Version]
- Pabinger, I.; van Es, N.; Heinze, G.; Posch, F.; Riedl, J.; Reitter, E.M.; Di Nisio, M.; Cesarman-Maus, G.; Kraaijpoel, N.; Zielinski, C.C.; et al. A clinical prediction model for cancer-associated venous thromboembolism: A development and validation study in two independent prospective cohorts. Lancet Haematol. 2018, 5, e289. [Google Scholar] [CrossRef]
- Zou, H.; Wen, C.; Peng, Z.; Shao, Y.; Hu, L.; Li, S.; Li, C.; Zhou, H.H. P4HB and PDIA3 are associated with tumor progression and therapeutic outcome of diffuse gliomas. Oncol. Rep. 2018, 39, 501–510. [Google Scholar] [CrossRef]
- Chen, F.; Zhao, Z.; Zhou, J.; Lu, Y.; Essex, D.W.; Wu, Y. Protein disulfide isomerase enhances tissue factor-dependent thrombin generation. Biochem. Biophys. Res. Commun. 2018, 501, 172–177. [Google Scholar] [CrossRef] [PubMed]
- Chen, V.M.; Ahamed, J.; Versteeg, H.H.; Berndt, M.C.; Ruf, W.; Hogg, P.J. Evidence for activation of tissue factor by an allosteric disulfide bond. Biochemistry 2006, 45, 12020–12028. [Google Scholar] [CrossRef]
- Langer, F.; Ruf, W. Synergies of phosphatidylserine and protein disulfide isomerase in tissue factor activation. Thromb. Haemost. 2014, 111, 590–597. [Google Scholar] [CrossRef] [Green Version]
- Stopa, J.D.; Neuberg, D.; Puligandla, M.; Furie, B.; Flaumenhaft, R.; Zwicker, J.I. Protein disulfide isomerase inhibition blocks thrombin generation in humans by interfering with platelet factor V activation. JCI Insight 2017, 2, e89373. [Google Scholar] [CrossRef] [PubMed]
- Giannakopoulos, B.; Gao, L.; Qi, M.; Wong, J.W.; Yu, D.M.; Vlachoyiannopoulos, P.G.; Moutsopoulos, H.M.; Atsumi, T.; Koike, T.; Hogg, P.; et al. Factor XI is a substrate for oxidoreductases: Enhanced activation of reduced FXI and its role in antiphospholipid syndrome thrombosis. J. Autoimmun. 2012, 39, 121–129. [Google Scholar] [CrossRef] [PubMed]
- Jurk, K.; Lahav, J.; Van Aken, H.; Brodde, M.F.; Nofer, J.R.; Kehrel, B.E. Extracellular protein disulfide isomerase regulates feedback activation of platelet thrombin generation via modulation of coagulation factor binding. J. Thromb. Haemost. 2011, 9, 2278–2290. [Google Scholar] [CrossRef] [PubMed]
- Wei, W.; Misra, S.; Cannon, M.V.; Yang, R.; Zhu, X.; Gilmore, R.; Zhu, M.; Zhang, B. Molecular mechanisms of missense mutations that generate ectopic N-glycosylation sites in coagulation factor VIII. Biochem. J. 2018, 475, 873–886. [Google Scholar] [CrossRef]
- Poothong, J.; Pottekat, A.; Siirin, M.; Campos, A.R.; Paton, A.W.; Paton, J.C.; Lagunas-Acosta, J.; Chen, Z.; Swift, M.; Volkmann, N.; et al. Factor VIII exhibits chaperone-dependent and glucose-regulated reversible amyloid formation in the endoplasmic reticulum. Blood 2020, 135, 1899–1911. [Google Scholar] [CrossRef] [PubMed]
- Du, L.M.; Nurden, P.; Nurden, A.T.; Nichols, T.C.; Bellinger, D.A.; Jensen, E.S.; Haberichter, S.L.; Merricks, E.; Raymer, R.A.; Fang, J.; et al. Platelet-Targeted gene therapy with human factor VIII establishes haemostasis in dogs with haemophilia A. Nat. Commun. 2013, 4, 2773. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Riedl, J.; Ay, C.; Pabinger, I. Platelets and hemophilia: A review of the literature. Thromb. Res. 2017, 155, 131–139. [Google Scholar] [CrossRef] [PubMed]
- Lyde, R.B.; Ahn, H.S.; Vo, K.K.; Jarocha, D.J.; Tkaczynski, J.; Treffeisen, E.; Sullivan, S.K.; Camire, R.M.; Sabatino, D.E.; French, D.L.; et al. Infused factor VIII-expressing platelets or megakaryocytes as a novel therapeutic strategy for hemophilia A. Blood Adv. 2019, 3, 1368–1378. [Google Scholar] [CrossRef] [PubMed]
- Vormittag, R.; Simanek, R.; Ay, C.; Dunkler, D.; Quehenberger, P.; Marosi, C.; Zielinski, C.; Pabinger, I. High factor VIII levels independently predict venous thromboembolism in cancer patients: The cancer and thrombosis study. Arterioscler. Thromb. Vasc. Biol. 2009, 29, 2176–2181. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Castellon Rubio, V.E.; Segura, P.P.; Munoz, A.; Farre, A.L.; Ruiz, L.C.; Lorente, J.A. High plasma levels of soluble P-Selectin and Factor VIII predict venous thromboembolism in non-small cell lung cancer patients: The Thrombo-Nsclc risk score. Thromb. Res. 2020, 196, 349–354. [Google Scholar] [CrossRef] [PubMed]
- Ay, C.; Vormittag, R.; Dunkler, D.; Simanek, R.; Chiriac, A.L.; Drach, J.; Quehenberger, P.; Wagner, O.; Zielinski, C.; Pabinger, I. D-dimer and prothrombin fragment 1 + 2 predict venous thromboembolism in patients with cancer: Results from the Vienna Cancer and Thrombosis Study. J. Clin. Oncol. 2009, 27, 4124–4129. [Google Scholar] [CrossRef]
- Tiedje, V.; Dunkler, D.; Ay, C.; Horvath, B.; Quehenberger, P.; Pabinger, M.; Zielinski, C.; Pabinger, I.; Mannhalter, C. The role of fibrinogen plasma levels, the −455 G > A fibrinogen and the factor XIII A subunit (FXIII-A) Val34Leu polymorphism in cancer-associated venous thrombosis. Thromb. Haemost. 2011, 106, 908–913. [Google Scholar] [CrossRef]
- Michelson, A.D.; Barnard, M.R.; Hechtman, H.B.; MacGregor, H.; Connolly, R.J.; Loscalzo, J.; Valeri, C.R. In vivo tracking of platelets: Circulating degranulated platelets rapidly lose surface P-selectin but continue to circulate and function. Proc. Natl. Acad. Sci. USA 1996, 93, 11877–11882. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Riedl, J.; Preusser, M.; Nazari, P.M.; Posch, F.; Panzer, S.; Marosi, C.; Birner, P.; Thaler, J.; Brostjan, C.; Lotsch, D.; et al. Podoplanin expression in primary brain tumors induces platelet aggregation and increases risk of venous thromboembolism. Blood 2017, 129, 1831–1839. [Google Scholar] [CrossRef] [Green Version]
- Al-Horani, R.A.; Kar, S. Factor XIIIa inhibitors as potential novel drugs for venous thromboembolism. Eur. J. Med. Chem. 2020, 200, 112442. [Google Scholar] [CrossRef]
- Byrnes, J.R.; Duval, C.; Wang, Y.; Hansen, C.E.; Ahn, B.; Mooberry, M.J.; Clark, M.A.; Johnsen, J.M.; Lord, S.T.; Lam, W.A.; et al. Factor XIIIa-dependent retention of red blood cells in clots is mediated by fibrin alpha-chain crosslinking. Blood 2015, 126, 1940–1948. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Muszbek, L.; Yee, V.C.; Hevessy, Z. Blood coagulation factor XIII: Structure and function. Thromb. Res. 1999, 94, 271–305. [Google Scholar] [CrossRef]
- Mitchell, J.L.; Lionikiene, A.S.; Fraser, S.R.; Whyte, C.S.; Booth, N.A.; Mutch, N.J. Functional factor XIII-A is exposed on the stimulated platelet surface. Blood 2014, 124, 3982–3990. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Byrnes, J.R.; Wolberg, A.S. Newly-Recognized roles of Factor XIII in thrombosis. Semin. Thromb. Hemost. 2016, 42, 445–454. [Google Scholar] [CrossRef] [Green Version]
- Muszbek, L.; Polgar, J.; Boda, Z. Platelet factor XIII becomes active without the release of activation peptide during platelet activation. Thromb. Haemost. 1993, 69, 282–285. [Google Scholar] [CrossRef] [PubMed]
- Komaromi, I.; Bagoly, Z.; Muszbek, L. Factor XIII: Novel structural and functional aspects. J. Thromb. Haemost. 2011, 9, 9–20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kool, R.O.; Kohler, H.P.; Coutinho, J.M.; Levi, M.; Coppens, M.; Meijers, J.C.M.; Schroeder, V. Coagulation factor XIII-A subunit and activation peptide levels in individuals with established symptomatic acute deep vein thrombosis. Thromb. Res. 2017, 159, 96–99. [Google Scholar] [CrossRef] [PubMed]
- Palacios-Acedo, A.L.; Mege, D.; Crescence, L.; Dignat-George, F.; Dubois, C.; Panicot-Dubois, L. Platelets, Thrombo-Inflammation, and cancer: Collaborating with the enemy. Front. Immunol. 2019, 10, 1805. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Palumbo, J.S.; Barney, K.A.; Blevins, E.A.; Shaw, M.A.; Mishra, A.; Flick, M.J.; Kombrinck, K.W.; Talmage, K.E.; Souri, M.; Ichinose, A.; et al. Factor XIII transglutaminase supports hematogenous tumor cell metastasis through a mechanism dependent on natural killer cell function. J. Thromb. Haemost. 2008, 6, 812–819. [Google Scholar] [CrossRef] [PubMed]
- Raval, J.S.; Berg, A.N.; Djokic, M.; Roth, C.G.; Rollins-Raval, M.A. Factor XIII subunit A immunohistochemical expression is associated with inferior outcomes in acute promyelocytic leukemia. Appl. Immunohistochem. Mol. Morphol. 2018, 26, 202–205. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.H.; Suh, I.B.; Lee, E.J.; Hur, G.Y.; Lee, S.Y.; Lee, S.Y.; Shin, C.; Shim, J.J.; In, K.H.; Kang, K.H.; et al. Relationships of coagulation factor XIII activity with cell-type and stage of non-small cell lung cancer. Yonsei Med. J. 2013, 54, 1394–1399. [Google Scholar] [CrossRef] [Green Version]
- Porrello, A.; Leslie, P.L.; Harrison, E.B.; Gorentla, B.K.; Kattula, S.; Ghosh, S.K.; Azam, S.H.; Holtzhausen, A.; Chao, Y.L.; Hayward, M.C.; et al. Factor XIIIA-expressing inflammatory monocytes promote lung squamous cancer through fibrin cross-linking. Nat. Commun. 2018, 9, 1988. [Google Scholar] [CrossRef] [Green Version]
- Urra, H.; Dufey, E.; Avril, T.; Chevet, E.; Hetz, C. Endoplasmic reticulum stress and the hallmarks of cancer. Trends Cancer 2016, 2, 252–262. [Google Scholar] [CrossRef]
- Muse, O.; Patell, R.; Peters, C.; Schulman, S.; Darzi, E.; Schuster, C.; Huang, L.; Mukherjee, S.; Muthuswamy, S.; Zwicker, J.I.; et al. The unfolded protein response causes prothrombotic transformation of pancreatic cancer linking tumor progression with cancer-associated thrombosis. Blood 2019, 134. [Google Scholar] [CrossRef]
- Zwicker, J.I.; Schlechter, B.L.; Stopa, J.D.; Liebman, H.A.; Aggarwal, A.; Puligandla, M.; Caughey, T.; Bauer, K.A.; Kuemmerle, N.; Wong, E.; et al. Targeting protein disulfide isomerase with the flavonoid isoquercetin to improve hypercoagulability in advanced cancer. JCI Insight 2019, 4. [Google Scholar] [CrossRef] [Green Version]
- Chang, I.J.; Byers, H.M.; Ng, B.G.; Merritt, J.L., 2nd; Gilmore, R.; Shrimal, S.; Wei, W.; Zhang, Y.; Blair, A.B.; Freeze, H.H.; et al. Factor VIII and vWF deficiency in STT3A-CDG. J. Inherit. Metab. Dis. 2019, 42, 325–332. [Google Scholar] [CrossRef] [PubMed]
- Dorner, A.J.; Wasley, L.C.; Kaufman, R.J. Increased synthesis of secreted proteins induces expression of glucose-regulated proteins in butyrate-treated Chinese hamster ovary cells. J. Biol. Chem. 1989, 264, 20602–20607. [Google Scholar] [CrossRef]
- Rietveld, I.M.; Lijfering, W.M.; le Cessie, S.; Bos, M.H.A.; Rosendaal, F.R.; Reitsma, P.H.; Cannegieter, S.C. High levels of coagulation factors and venous thrombosis risk: Strongest association for factor VIII and von Willebrand factor. J. Thromb. Haemost. 2019, 17, 99–109. [Google Scholar] [CrossRef] [Green Version]
- Fahs, S.A.; Hille, M.T.; Shi, Q.; Weiler, H.; Montgomery, R.R. A conditional knockout mouse model reveals endothelial cells as the principal and possibly exclusive source of plasma factor VIII. Blood 2014, 123, 3706–3713. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Paoli, S.H.; Tegegn, T.Z.; Elhelu, O.K.; Strader, M.B.; Patel, M.; Diduch, L.L.; Tarandovskiy, I.D.; Wu, Y.; Zheng, J.; Ovanesov, M.V.; et al. Dissecting the biochemical architecture and morphological release pathways of the human platelet extracellular vesiculome. Cell. Mol. Life Sci. 2018, 75, 3781–3801. [Google Scholar] [CrossRef] [PubMed]
- Veitinger, M.; Umlauf, E.; Baumgartner, R.; Badrnya, S.; Porter, J.; Lamont, J.; Gerner, C.; Gruber, C.; Oehler, R.; Zellner, M. A combined proteomic and genetic analysis of the highly variable platelet proteome: From plasmatic proteins and SNPs. J. Proteom. 2012, 75, 5848–5860. [Google Scholar] [CrossRef] [PubMed]
- Campello, E.; Spiezia, L.; Radu, C.M.; Bulato, C.; Castelli, M.; Gavasso, S.; Simioni, P. Endothelial, platelet, and tissue factor-bearing microparticles in cancer patients with and without venous thromboembolism. Thromb. Res. 2011, 127, 473–477. [Google Scholar] [CrossRef] [PubMed]
- Chaturvedi, S.; Alluri, R.; McCrae, K.R. Extracellular vesicles in the antiphospholipid syndrome. Semin. Thromb. Hemost. 2018, 44, 493–504. [Google Scholar] [CrossRef]
- Grilz, E.; Mauracher, L.M.; Posch, F.; Konigsbrugge, O.; Zochbauer-Muller, S.; Marosi, C.; Lang, I.; Pabinger, I.; Ay, C. Citrullinated histone H3, a biomarker for neutrophil extracellular trap formation, predicts the risk of mortality in patients with cancer. Br. J. Haematol. 2019, 186, 311–320. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Westermeier, R. 2D gel-based proteomics: There’s life in the old dog yet. Arch. Physiol. Biochem. 2016, 122, 236–237. [Google Scholar] [CrossRef] [PubMed]







| Characteristic | Healthy Controls (n = 41) | Healthy Controls for Brain Cancer (n = 22) | Healthy Controls for Lung Cancer (n = 19) | Brain Cancer (n = 22) | Lung Cancer (n = 19) |
|---|---|---|---|---|---|
| Median age at study entry, y (IQR) | 57 (48–62) | 55 (42–60) | 61 (56–63) | 56 (45–63) | 62 (58–67) |
| Female, n (%) | 16 (39) | 9 (41) | 7 (37) | 9 (41) | 7 (37) |
| Median laboratory values (IQR) | |||||
| Leukocyte (G/L) | 5.9 (5.1–7.1) | 5.7 (4.9–7.8) | 5.9 (5.2–6.9) $$$ | 7.1 (5.4–11.8) | 9.8 (7.6–11.1) |
| Hemoglobin (G/dL) | 14.5 (13.2–15.4) | 14.8 (13.1–15.6) | 14.3 (13–15.3) $ | 14.3 (13.3–15.2) | 12.9 (12.2–14) |
| Platelet count (G/L) | 235 (216–286) | 235 (214.5–308.5) | 230 (225–286) $ | 239 (202–297) | 297 (276–350) |
| MPV (fl) | 10.8 (10–11.4) | 10.9 (10.1–11.4) # | 10.8 (9.9–11.5) $ | 10 (9.7–10.7) | 10.1 (9.2–10.8) |
| Neutrophils (%) | 58 (51–64.5) | 60 (48–61.8) ### | 53 (54–68) $$$ | 73 (56.7–76.1) | 82.4 (76–87.7) |
| CRP (mg/dL) | 0.14 (0.06–0.25) | 0.16 (0.06–0.23) | 0.09 (0.07–0.32) $$$ | 0.14 (0.06–0.27) | 1.16 (0.65–2.8) |
| aPTT (s) | 35 (32.1–37.7) | 33 (33.1–38.1) # | 36 (31.3–35.8) | 34.1 (29.4–36) | 33.5 (31.8–40.7) |
| Fibrinogen (mg/dL) | 303 (259–337) | 313 (250.5–319. 5) ## | 282 (280–353) $$$ | 319 (305–375) | 509 (427–639) |
| Prothrombinfragment (pmol/L) | 180 (127–249) | 192 (118.5–297) | 165 (148.5–225.8) | 165 (109–199) | 235 (154–349) |
| FVIII activity (%) | 147 (107–173) | 165 (103.5–154) ### | 135 (117–184) $$ | 208 (167–265) | 237 (166–369) |
| Antithrombin III (%) | 103 (97–108) | 105 (96.5–108.5) ### | 100 (101–108) | 122 (111–133) | 105 (85–110) |
| PAI (IU/mL) | 1.2 (0.5–5) | 1.1 (0.49–6) | 1.5 (0.49–4.2) | 2.1 (0.53–5.7) | 1.4 (0.9–6.6) |
| Plasma FXIII activity (%) | 123 (111–140) | 122.5 (104.5–135.4) # | 120.4 (115.5–151) | 106.8 (91.1–111) | 120.2 (95.6–139.4) |
| D-dimer (µg/mL) | 0.32 (0.27–0.42) | 0.33 (0.27–0.41) # | 0.3 (0.27–0.42) $$$ | 0.74 (0.33–0.93) | 1.79 (1.13–4.15) |
| VTE during follow up, n (%) | n.a. | n.a | n.a | 2 (9.1) | 2 (10.5) |
| PE | n.a | n.a | n.a | 1 (5) | 2 (10.5) |
| DVT | n.a | n.a | n.a | 1 (5) | n.a. |
| Deaths during follow up, n (%) | n.a | n.a | n.a | 8 (36.4) | 11 (57.9) |
| Brain Cancer Patients/ Healthy Controls | Lung Cancer Patients/ Healthy Controls | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Spot Number | Protein Name | Gene Name | Isoelectric Point (pI) | MW (Da) | One-Way ANOVA (Adjusted) | Average FC | p-Value (Unadjusted) | p-Value (Adjusted) | Average FC | p-Value (Unadjusted) | p-Value (Adjusted) |
| 1 | Albumin | ALB | 6.00 | 69,367 | 0.0224 | 0.84 | 0.0114 | 0.1136 | 0.85 | 0.0281 | 0.0388 |
| 2 | Basement-membrane protein 40 (SPARC; Osteonectin) | SPARC | 4.75 | 42,014 | 0.0488 | 0.96 | 0.6911 | 0.8639 | 0.62 | 0.0001 | 0.0013 |
| 3 | Calreticulin | CALR | 4.29 | 67,014 | 0.0127 | 1.06 | 0.1427 | 0.4926 | 1.10 | 0.0315 | 0.0420 |
| 4 | 4.29 | 65,300 | 0.0224 | 1.11 | 0.0254 | 0.1451 | 1.12 | 0.0524 | 0.0655 | ||
| 5 | 4.29 | 64,385 | 0.0264 | 1.09 | 0.0301 | 0.1505 | 1.11 | 0.0616 | 0.0747 | ||
| 6 | 4.29 | 63,493 | 0.0188 | 1.13 | 0.0069 | 0.1053 | 1.11 | 0.0156 | 0.0240 | ||
| 7 | 4.29 | 61,919 | 0.0433 | 1.14 | 0.0191 | 0.1273 | 1.12 | 0.0123 | 0.0223 | ||
| 8 | 4.29 | 60,228 | 0.0482 | 1.18 | 0.0074 | 0.1053 | 1.16 | 0.0515 | 0.0655 | ||
| 9 | 4.29 | 56,324 | 0.0224 | 1.14 | 0.0079 | 0.1053 | 1.12 | 0.0054 | 0.0135 | ||
| 10 | 4.29 | 57,598 | 0.0500 | 1.19 | 0.0142 | 0.1136 | 1.12 | 0.0144 | 0.0238 | ||
| 11 | Coagulation factor XIII A chain | F13A1 | 5.00 | 55,000 | 0.0025 | 0.95 | 0.4346 | 0.7243 | 1.57 | 0.0016 | 0.0080 |
| 12 | 5.75 | 83,267 | 0.0500 | 0.90 | 0.2608 | 0.5611 | 1.21 | 0.0102 | 0.0194 | ||
| 13 | Endoplasmic reticulum resident protein 29 | ERP29 | 5.95 | 27,022 | 0.0188 | 1.06 | 0.2242 | 0.501 | 1.20 | 0.013 | 0.0212 |
| 14 | Endoplasmic reticulum chaperone BiP | HSPA5 | 5.06 | 73,539 | 0.0488 | 0.99 | 0.8875 | 0.9342 | 1.07 | 0.1212 | 0.1347 |
| 15 | 5.08 | 73,539 | 0.0224 | 1.06 | 0.3828 | 0.6960 | 1.15 | 0.0098 | 0.0194 | ||
| 16 | Eukaryotic translation initiation factor 2 subunit 1 | EIF2S1 | 5.08 | 43,211 | 0.0188 | 0.99 | 0.8726 | 0.9342 | 0.83 | 0.0018 | 0.0080 |
| 17 | Fermitin family homolog 3 | FERMT3 | 6.00 | 39,072 | 0.0019 | 0.94 | 0.5676 | 0.8148 | 1.11 | 0.1191 | 0.1347 |
| 18 | Fibrinogen gamma chain | FGG | 5.74 | 55,146 | 0.0467 | 1.03 | 0.4800 | 0.7385 | 1.18 | 0.0022 | 0.0080 |
| 19 | 5.58 | 56,048 | 0.0488 | 1.45 | 0.2812 | 0.5624 | 1.47 | 0.0032 | 0.0091 | ||
| 20 | Gelsolin | GSN | 5.70 | 81,086 | 0.0188 | 1.07 | 0.0989 | 0.3956 | 0.92 | 0.0221 | 0.0316 |
| 21 | Integrin alpha-IIb | ITGA2B | 4.80 | 88,938 | 0.0467 | 0.94 | 0.1933 | 0.5155 | 0.88 | 0.0052 | 0.0135 |
| 22 | 4.92 | 88,938 | 0.0096 | 0.93 | 0.1562 | 0.4926 | 0.86 | 0.0001 | 0.0013 | ||
| 23 | 4.95 | 88,938 | 0.0224 | 0.91 | 0.0641 | 0.2849 | 0.86 | 0.0020 | 0.0080 | ||
| 24 | Integrin beta-3 | ITGB3 | 4.68 | 75,464 | 0.0127 | 1.00 | 0.9763 | 0.9763 | 0.82 | 0.0008 | 0.0053 |
| 25 | 4.70 | 75,464 | 0.0390 | 0.95 | 0.2597 | 0.5611 | 0.87 | 0.0061 | 0.0144 | ||
| 26 | 4.73 | 75,464 | 0.0467 | 0.96 | 0.3805 | 0.6960 | 0.88 | 0.0094 | 0.0194 | ||
| 27 | 5.63 | 75,464 | 0.0046 | 0.90 | 0.2665 | 0.5611 | 0.63 | 0.0005 | 0.0040 | ||
| 28 | Protein disulfide-isomerease | P4HB | 4.78 | 61,026 | 0.0127 | 1.01 | 0.8415 | 0.9342 | 1.11 | 0.0016 | 0.0080 |
| 29 | 4.80 | 61,026 | 0.0019 | 0.98 | 0.5906 | 0.8148 | 1.11 | 0.0211 | 0.0313 | ||
| 30 | Protein disulfide-isomerase A3 | PDIA3 | 5.81 | 60,726 | 0.0467 | 1.02 | 0.4246 | 0.7243 | 1.03 | 0.2144 | 0.2199 |
| 31 | 5.81 | 58,443 | 0.0417 | 1.04 | 0.1645 | 0.4926 | 1.06 | 0.1529 | 0.1653 | ||
| 32 | Rab GDP dissociation inhibitor alpha | GDI1 | 4.95 | 62,247 | 0.0019 | 1.02 | 0.6780 | 0.8639 | 1.31 | 0.0002 | 0.0020 |
| 33 | Serotransferrin | TF | 6.62 | 76,915 | 0.0272 | 0.95 | 0.4797 | 0.7385 | 0.79 | 0.0149 | 0.0238 |
| 34 | Talin-1 | TLN1 | 5.31 | 75,981 | 0.0096 | 0.97 | 0.7319 | 0.8872 | 1.45 | 0.0072 | 0.0160 |
| 35 | 14-3-3 protein epsilon | YWHAE | 4.60 | 28,953 | 0.0420 | 1.01 | 0.5907 | 0.8148 | 0.98 | 0.3459 | 0.3459 |
| 36 | 14-3-3 protein zeta/delta | YWHAZ | 4.69 | 26,607 | 0.0420 | 0.99 | 0.8331 | 0.9342 | 0.96 | 0.0673 | 0.0792 |
| Lung Cancer Patients/ Matched Controls | Brain Cancer Patients/ Matched Controls | Correlation: Abundance vs. Enzymatic Activity | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Spot Number of F13A1 Proteoforms | Isoelectric Point (pI) | MW (kDa) | ANOVA (Adjusted) | Average FC | p-Value (Unadjusted) | Average FC | p-Value (Unadjusted) | corr. of F13A1 Abundance vs. Activity→Rho | p-Value (Unadjusted) |
| 12c | 5.85 | 83 | 0.998 | 0.77 | 0.2652 | 1.24 | 0.372 | −0.29 | 0.062 |
| 12b | 5.75 | 83 | 0.410 | 1.40 | 0.0019 | 0.96 | 0.743 | 0.43 | 0.004 |
| 12 | 5.60 | 83 | 0.071 | 1.21 | 0.0102 | 0.90 | 0.261 | 0.39 | 0.009 |
| 12a | 5.65 | 83 | 0.198 | 1.33 | 0.0369 | 0.82 | 0.231 | 0.50 | 0.001 |
| 12d | 6.05 | 79 | 0.868 | 1.26 | 0.3055 | 1.05 | 0.832 | 0.04 | 0.807 |
| 11 | 4.95 | 55 | 0.006 | 1.57 | 0.0016 | 0.95 | 0.433 | 0.14 | 0.367 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ercan, H.; Mauracher, L.-M.; Grilz, E.; Hell, L.; Hellinger, R.; Schmid, J.A.; Moik, F.; Ay, C.; Pabinger, I.; Zellner, M. Alterations of the Platelet Proteome in Lung Cancer: Accelerated F13A1 and ER Processing as New Actors in Hypercoagulability. Cancers 2021, 13, 2260. https://doi.org/10.3390/cancers13092260
Ercan H, Mauracher L-M, Grilz E, Hell L, Hellinger R, Schmid JA, Moik F, Ay C, Pabinger I, Zellner M. Alterations of the Platelet Proteome in Lung Cancer: Accelerated F13A1 and ER Processing as New Actors in Hypercoagulability. Cancers. 2021; 13(9):2260. https://doi.org/10.3390/cancers13092260
Chicago/Turabian StyleErcan, Huriye, Lisa-Marie Mauracher, Ella Grilz, Lena Hell, Roland Hellinger, Johannes A. Schmid, Florian Moik, Cihan Ay, Ingrid Pabinger, and Maria Zellner. 2021. "Alterations of the Platelet Proteome in Lung Cancer: Accelerated F13A1 and ER Processing as New Actors in Hypercoagulability" Cancers 13, no. 9: 2260. https://doi.org/10.3390/cancers13092260
APA StyleErcan, H., Mauracher, L. -M., Grilz, E., Hell, L., Hellinger, R., Schmid, J. A., Moik, F., Ay, C., Pabinger, I., & Zellner, M. (2021). Alterations of the Platelet Proteome in Lung Cancer: Accelerated F13A1 and ER Processing as New Actors in Hypercoagulability. Cancers, 13(9), 2260. https://doi.org/10.3390/cancers13092260