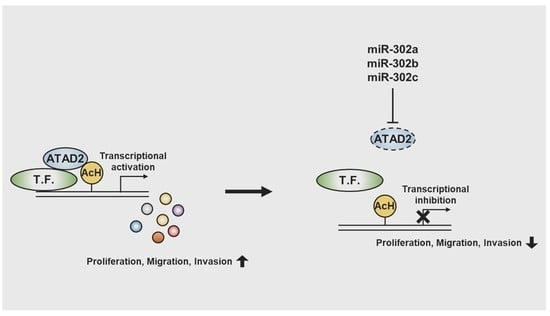
miR-302 Suppresses the Proliferation, Migration, and Invasion of Breast Cancer Cells by Downregulating ATAD2
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. Preparation of miRNA, DNA Constructs, and Transfection
2.3. miRNA Prediction
2.4. Immunohistochemistry (IHC)
2.5. Reverse Transcription-Polymerase Chain Reaction (RT-PCR) and Quantitative RT-PCR (qRT-PCR)
2.6. Western Blotting
2.7. Cell Proliferation Assay
2.8. Cell Migration and Invasion Assays
2.9. Luciferase Reporter Assay
2.10. Animal Experiments
2.11. Statistical Analysis
3. Results
3.1. ATAD2 Expression Is Increased in Breast Cancer Tissues and Cell Lines
3.2. ATAD2 Promotes Breast Cancer Cell Proliferation, Migration, and Invasion
3.3. miR-302 Family Inhibits Breast Cancer Cell Migration and Invasion by Directly Targeting ATAD2
3.4. miR-302 Suppresses Tumor Growth In Vivo by Targeting ATAD2
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hussain, M.; Zhou, Y.; Song, Y.; Hameed, H.M.A.; Jiang, H.; Tu, Y.; Zhang, J. ATAD2 in cancer: A pharmacologically challenging but tractable target. Expert Opin. Targets 2018, 22, 85–96. [Google Scholar] [CrossRef] [PubMed]
- Boussouar, F.; Jamshidikia, M.; Morozumi, Y.; Rousseaux, S.; Khochbin, S. Malignant genome reprogramming by ATAD2. Biochim. Biophys. Acta 2013, 1829, 1010–1014. [Google Scholar] [CrossRef]
- Nayak, A.; Dutta, M.; Roychowdhury, A. Emerging oncogene ATAD2: Signaling cascades and therapeutic initiatives. Life Sci. 2021, 276, 119322. [Google Scholar] [CrossRef] [PubMed]
- Morozumi, Y.; Boussouar, F.; Tan, M.; Chaikuad, A.; Jamshidikia, M.; Colak, G.; He, H.; Nie, L.; Petosa, C.; de Dieuleveult, M.; et al. Atad2 is a generalist facilitator of chromatin dynamics in embryonic stem cells. J. Mol. Cell Biol. 2016, 8, 349–362. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Li, S.; Cheng, K.W.; Chou, T.F. AAA ATPases as therapeutic targets: Structure, functions, and small-molecule inhibitors. Eur. J. Med. Chem. 2021, 219, 113446. [Google Scholar] [CrossRef]
- Zou, J.X.; Revenko, A.S.; Li, L.B.; Gemo, A.T.; Chen, H.W. ANCCA, an estrogen-regulated AAA+ ATPase coactivator for ERalpha, is required for coregulator occupancy and chromatin modification. Proc. Natl. Acad. Sci. USA 2007, 104, 18067–18072. [Google Scholar] [CrossRef]
- Zou, J.X.; Guo, L.; Revenko, A.S.; Tepper, C.G.; Gemo, A.T.; Kung, H.J.; Chen, H.W. Androgen-induced coactivator ANCCA mediates specific androgen receptor signaling in prostate cancer. Cancer Res. 2009, 69, 3339–3346. [Google Scholar] [CrossRef]
- Revenko, A.S.; Kalashnikova, E.V.; Gemo, A.T.; Zou, J.X.; Chen, H.W. Chromatin loading of E2F-MLL complex by cancer-associated coregulator ANCCA via reading a specific histone mark. Mol. Cell Biol. 2010, 30, 5260–5272. [Google Scholar] [CrossRef]
- Ciró, M.; Prosperini, E.; Quarto, M.; Grazini, U.; Walfridsson, J.; McBlane, F.; Nucifero, P.; Pacchiana, G.; Capra, M.; Christensen, J.; et al. ATAD2 is a novel cofactor for MYC, overexpressed and amplified in aggressive tumors. Cancer Res. 2009, 69, 8491–8498. [Google Scholar] [CrossRef]
- Luo, Y.; Ye, G.Y.; Qin, S.L.; Yu, M.H.; Mu, Y.F.; Zhong, M. ATAD2 Overexpression Identifies Colorectal Cancer Patients with Poor Prognosis and Drives Proliferation of Cancer Cells. Gastroenterol. Res. Pract. 2015, 2015, 936564. [Google Scholar] [CrossRef] [Green Version]
- Zhang, M.; Zhang, C.; Du, W.; Yang, X.; Chen, Z. ATAD2 is overexpressed in gastric cancer and serves as an independent poor prognostic biomarker. Clin. Transl. Oncol. 2016, 18, 776–781. [Google Scholar] [CrossRef]
- Nayak, A.; Roy, A.D.; Rout, N.; Singh, S.P.; Bhattacharyya, A.; Roychowdhury, A. HIF1alpha-dependent upregulation of ATAD2 promotes proliferation and migration of stomach cancer cells in response to hypoxia. Biochem. Biophys. Res. Commun. 2020, 523, 916–923. [Google Scholar] [CrossRef] [PubMed]
- Caron, C.; Lestrat, C.; Marsal, S.; Escoffier, E.; Curtet, S.; Virolle, V.; Barbry, P.; Debernardi, A.; Brambilla, C.; Brambilla, E.; et al. Functional characterization of ATAD2 as a new cancer/testis factor and a predictor of poor prognosis in breast and lung cancers. Oncogene 2010, 29, 5171–5181. [Google Scholar] [CrossRef]
- Zhang, Y.; Sun, Y.; Li, Y.; Fang, Z.; Wang, R.; Pan, Y.; Hu, H.; Luo, X.; Ye, T.; Li, H.; et al. ANCCA protein expression is a novel independent poor prognostic marker in surgically resected lung adenocarcinoma. Ann. Surg. Oncol. 2013, 20 (Suppl. 3), S577–S582. [Google Scholar] [CrossRef] [PubMed]
- Kalashnikova, E.V.; Revenko, A.S.; Gemo, A.T.; Andrews, N.P.; Tepper, C.G.; Zou, J.X.; Cardiff, R.D.; Borowsky, A.D.; Chen, H.W. ANCCA/ATAD2 overexpression identifies breast cancer patients with poor prognosis, acting to drive proliferation and survival of triple-negative cells through control of B-Myb and EZH2. Cancer Res. 2010, 70, 9402–9412. [Google Scholar] [CrossRef] [PubMed]
- De Angelis, P.M.; Svendsrud, D.H.; Kravik, K.L.; Stokke, T. Cellular response to 5-fluorouracil (5-FU) in 5-FU-resistant colon cancer cell lines during treatment and recovery. Mol. Cancer 2006, 5, 20. [Google Scholar] [CrossRef]
- Murakami, H.; Ito, S.; Tanaka, H.; Kondo, E.; Kodera, Y.; Nakanishi, H. Establishment of new intraperitoneal paclitaxel-resistant gastric cancer cell lines and comprehensive gene expression analysis. Anticancer Res. 2013, 33, 4299–4307. [Google Scholar]
- Zou, J.X.; Duan, Z.; Wang, J.; Sokolov, A.; Xu, J.; Chen, C.Z.; Li, J.J.; Chen, H.W. Kinesin family deregulation coordinated by bromodomain protein ANCCA and histone methyltransferase MLL for breast cancer cell growth, survival, and tamoxifen resistance. Mol. Cancer Res. 2014, 12, 539–549. [Google Scholar] [CrossRef]
- Volovat, S.R.; Volovat, C.; Hordila, I.; Hordila, D.A.; Mirestean, C.C.; Miron, O.T.; Lungulescu, C.; Scripcariu, D.V.; Stolniceanu, C.R.; Konsoulova-Kirova, A.A.; et al. MiRNA and LncRNA as Potential Biomarkers in Triple-Negative Breast Cancer: A Review. Front. Oncol. 2020, 10, 526850. [Google Scholar] [CrossRef]
- Ruffo, P.; Strafella, C.; Cascella, R.; Caputo, V.; Conforti, F.L.; Andò, S.; Giardina, E. Deregulation of ncRNA in Neurodegenerative Disease: Focus on circRNA, lncRNA and miRNA in Amyotrophic Lateral Sclerosis. Front. Genet. 2021, 12, 784996. [Google Scholar] [CrossRef]
- Mirzaei, S.; Zarrabi, A.; Hashemi, F.; Zabolian, A.; Saleki, H.; Ranjbar, A.; Seyed Saleh, S.H.; Bagherian, M.; Sharifzadeh, S.O.; Hushmandi, K.; et al. Regulation of Nuclear Factor-KappaB (NF-κB) signaling pathway by non-coding RNAs in cancer: Inhibiting or promoting carcinogenesis? Cancer Lett. 2021, 509, 63–80. [Google Scholar] [CrossRef] [PubMed]
- Wong, A.C.H.; Rasko, J.E.J. Splice and Dice: Intronic microRNAs, Splicing and Cancer. Biomedicines 2021, 9, 1268. [Google Scholar] [CrossRef] [PubMed]
- Gareev, I.; Beylerli, O.; Liang, Y.; Xiang, H.; Liu, C.; Xu, X.; Yuan, C.; Ahmad, A.; Yang, G. The Role of MicroRNAs in Therapeutic Resistance of Malignant Primary Brain Tumors. Front Cell Dev Biol. 2021, 9, 740303. [Google Scholar] [CrossRef] [PubMed]
- Hussen, B.M.; Abdullah, S.T.; Rasul, M.F.; Salihi, A.; Ghafouri-Fard, S.; Hidayat, H.J.; Taheri, M. MicroRNAs: Important Players in Breast Cancer Angiogenesis and Therapeutic Targets. Front Mol Biosci. 2021, 8, 764025. [Google Scholar] [CrossRef]
- Balkrishna, A.; Mittal, R.; Arya, V. Unveiling Novel Therapeutic Drug Targets and Prognostic Markers of Triple Negative Breast Cancer. Curr. Cancer Drug Targets 2021, 21, 907–918. [Google Scholar] [CrossRef]
- Liu, H.; Wen, Q.; Yan., S.; Zeng., W.; Zou., Y.; Liu., Q.; Zhang., G.; Zou., J.; Zou., X. Tumor-Promoting ATAD2 and Its Preclinical Challenges. Biomolecules 2022, 12, 1040. [Google Scholar] [CrossRef]
- Hsia, E.Y.; Kalashnikova, E.V.; Revenko, A.S.; Zou, J.X.; Borowsky, A.D.; Chen, H.W. Deregulated E2F and the AAA+ coregulator ANCCA drive proto-oncogene ACTR/AIB1 overexpression in breast cancer. Mol. Cancer Res. 2010, 8, 183–193. [Google Scholar] [CrossRef]
- Bartel, D.P. Metazoan MicroRNAs. Cell 2018, 173, 20–51. [Google Scholar] [CrossRef]
- Fu, Z.; Wang, L.; Li, S.; Chen, F.; Au-Yeung, K.K.; Shi, C. MicroRNA as an Important Target for Anticancer Drug Development. Front. Pharmacol. 2021, 12, 736323. [Google Scholar] [CrossRef]
- Hong, S.; Bi, M.; Chen, S.; Zhao, P.; Li, B.; Sun, D.; Tai, J. MicroRNA-520f suppresses growth of gastric carcinoma cells by target ATPase family AAA domain-containing protein 2 (ATAD2). Neoplasma 2016, 63, 873–879. [Google Scholar] [CrossRef]
- Wu, G.; Liu, H.; He, H.; Wang, Y.; Lu, X.; Yu, Y.; Xia, S.; Meng, X.; Liu, Y. miR-372 down-regulates the oncogene ATAD2 to influence hepatocellular carcinoma proliferation and metastasis. BMC Cancer 2014, 14, 107. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guan, X.; Zong, Z.H.; Chen, S.; Sang, X.B.; Wu, D.D.; Wang, L.L.; Liu, Y.; Zhao, Y. The role of miR-372 in ovarian carcinoma cell proliferation. Gene 2017, 624, 14–20. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Han, M.; Zhang, C. Overexpression of microRNA-186 inhibits angiogenesis in retinoblastoma via the Hedgehog signaling pathway by targeting ATAD2. J. Cell Physiol. 2019, 234, 19059–19072. [Google Scholar] [CrossRef] [PubMed]
- Wang, A.Q.; Lv, M.; Xu, Y.H.; Xie, P.M.; Dong, Y.Y. MiR-200b-5p inhibits proliferation of ovarian cancer cells by targeting ATAD2 and regulating PI3K/AKT signaling pathway. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 9860–9868. [Google Scholar]
- Ge, T.; Liu, T.; Guo, L.; Chen, Z.; Lou, G. MicroRNA-302 represses epithelial-mesenchymal transition and cisplatin resistance by regulating ATAD2 in ovarian carcinoma. Exp. Cell Res. 2020, 396, 112241. [Google Scholar] [CrossRef]
- Ramezankhani, B.; Taha, M.F.; Javeri, A. Vitamin C counteracts miR-302/367-induced reprogramming of human breast cancer cells and restores their invasive and proliferative capacity. J. Cell Physiol. 2019, 234, 2672–2682. [Google Scholar] [CrossRef]
- Hoseinbeyki, M.; Taha, M.F.; Javeri, A. miR-16 enhances miR-302/367-induced reprogramming and tumor suppression in breast cancer cells. IUBMB Life 2020, 72, 1075–1086. [Google Scholar] [CrossRef]
- Zhao, L.; Wang, Y.; Jiang, L.; He, M.; Bai, X.; Yu, L.; Wei, M. MiR-302a/b/c/d cooperatively sensitizes breast cancer cells to adriamycin via suppressing P-glycoprotein(P-gp) by targeting MAP/ERK kinase kinase 1 (MEKK1). J. Exp. Clin. Cancer Res. 2016, 35, 25. [Google Scholar] [CrossRef]
- Wang, Y.; Zhao, L.; Xiao, Q.; Jiang, L.; He, M.; Bai, X.; Ma, M.; Jiao, X.; Wei, M. miR-302a/b/c/d cooperatively inhibit BCRP expression to increase drug sensitivity in breast cancer cells. Gynecol. Oncol. 2016, 141, 592–601. [Google Scholar] [CrossRef]






Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hwang, Y.S.; Park, E.S.; Oh, B.M.; Uhm, T.G.; Yoon, S.R.; Park, J.-L.; Cho, H.J.; Lee, H.G. miR-302 Suppresses the Proliferation, Migration, and Invasion of Breast Cancer Cells by Downregulating ATAD2. Cancers 2022, 14, 4345. https://doi.org/10.3390/cancers14184345
Hwang YS, Park ES, Oh BM, Uhm TG, Yoon SR, Park J-L, Cho HJ, Lee HG. miR-302 Suppresses the Proliferation, Migration, and Invasion of Breast Cancer Cells by Downregulating ATAD2. Cancers. 2022; 14(18):4345. https://doi.org/10.3390/cancers14184345
Chicago/Turabian StyleHwang, Yo Sep, Eun Sun Park, Byung Moo Oh, Tae Gi Uhm, Suk Ran Yoon, Jong-Lyul Park, Hee Jun Cho, and Hee Gu Lee. 2022. "miR-302 Suppresses the Proliferation, Migration, and Invasion of Breast Cancer Cells by Downregulating ATAD2" Cancers 14, no. 18: 4345. https://doi.org/10.3390/cancers14184345
APA StyleHwang, Y. S., Park, E. S., Oh, B. M., Uhm, T. G., Yoon, S. R., Park, J. -L., Cho, H. J., & Lee, H. G. (2022). miR-302 Suppresses the Proliferation, Migration, and Invasion of Breast Cancer Cells by Downregulating ATAD2. Cancers, 14(18), 4345. https://doi.org/10.3390/cancers14184345