Enhanced Antitumoral Activity of Encapsulated BET Inhibitors When Combined with PARP Inhibitors for the Treatment of Triple-Negative Breast and Ovarian Cancers
Abstract
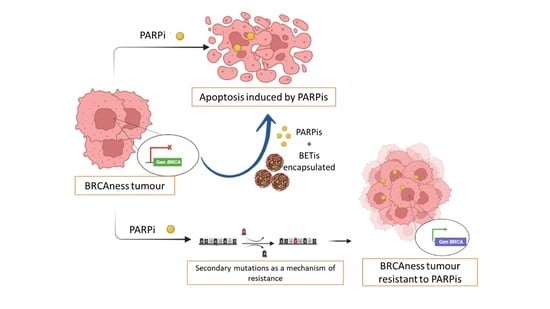
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Liposomal Formulation
2.3. JQ1-LP Polymeric Nanoparticle Formulation
2.4. Characterization of Formulations
2.5. Efficiency and Loading Efficiencies
2.6. Stability of NPs
2.7. In Vitro Drug-Release Studies
2.8. Cell Culture and Drug Compounds
2.9. MTT Proliferation Assay
2.10. EC50 Values
2.11. Cell Cycle and Apoptosis Assay
2.12. Synergy Studies
2.13. Statistical Analyses
3. Results
3.1. Formulation, Characterization, Releases Profiles and Stability of Nanodevices
3.2. Antitumour Effects of JQ1 and JQ1-Loaded Nanocarriers on Cell Proliferation
3.3. Effect on Proliferation of Non-Loaded Nanocarriers
3.4. Synergistic Interaction between JQ1, JQ1-LP or JQ1-NPs and Olaparib in Representative Models of Triple-Negative Breast and Ovarian Cancers
3.5. Free JQ1 and Encapsulated Formulations of JQ1 Induce G0/G1 Arrest
3.6. Cell Death Is Highly Induced with the Encapsulated Formulations of JQ1 in Combination with Olaparib
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Schettini, F.; Giudici, F.; Bernocchi, O.; Sirico, M.; Corona, S.P.; Giuliano, M.; Locci, M.; Paris, I.; Scambia, G.; Placido, S.D.; et al. Poly (ADP-ribose) polymerase inhibitors in solid tumours: Systematic review and meta-analysis. Eur. J. Cancer 2021, 149, 134–152. [Google Scholar] [CrossRef] [PubMed]
- Cortesi, L.; Rugo, H.S.; Jackisch, C. An Overview of PARP Inhibitors for the Treatment of Breast Cancer. Targ. Oncol. 2021, 16, 255–282. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Liu, Z.-Y.; Wu, N.; Chen, Y.-C.; Cheng, Q.; Wang, J. PARP inhibitor resistance: The underlying mechanisms and clinical implications. Mol. Cancer 2020, 19, 107. [Google Scholar] [CrossRef] [PubMed]
- Patel, M.; Nowsheen, S.; Maraboyina, S.; Xia, F. The role of poly(ADP-ribose) polymerase inhibitors in the treatment of cancer and methods to overcome resistance: A review. Cell Biosci. 2020, 10, 35. [Google Scholar] [CrossRef]
- Han, Y.; Yu, X.; Li, S.; Tian, Y.; Liu, C. New Perspectives for Resistance to PARP Inhibitors in Triple-Negative Breast Cancer. Front. Oncol. 2020, 10, 2456. [Google Scholar] [CrossRef]
- Raimundo, L.; Calheiros, J.; Saraiva, L. Exploiting DNA Damage Repair in Precision Cancer Therapy: BRCA1 as a Prime Therapeutic Target. Cancers 2021, 13, 3438. [Google Scholar] [CrossRef]
- Yi, M.; Dong, B.; Qin, S.; Chu, Q.; Wu, K.; Luo, S. Advances and perspectives of PARP inhibitors. Exp. Hematol. Oncol. 2019, 8, 29. [Google Scholar] [CrossRef]
- Bushweller, J.H. Targeting transcription factors in cancer—From undruggable to reality. Nat. Rev. Cancer 2019, 19, 611–624. [Google Scholar] [CrossRef]
- Zhang, W.; Prakash, C.; Sum, C.; Gong, Y.; Li, Y.; Kwok, J.J.T.; Thiessen, N.; Pettersson, S.; Jones, S.J.M.; Knapp, S.; et al. Bromodomain-containing Protein 4 (BRD4) Regulates RNA Polymerase II Serine 2 Phosphorylation in Human CD4+ T Cells. J. Biol. Chem. 2012, 287, 43137–43155. [Google Scholar] [CrossRef]
- Alqahtani, A.; Choucair, K.; Ashraf, M.; Hammouda, D.M.; Alloghbi, A.; Khan, T.; Senzer, N.; Nemunaitis, J. Bromodomain and extra-terminal motif inhibitors: A review of preclinical and clinical advances in cancer therapy. Future Sci. OA 2019, 5, FSO372. [Google Scholar] [CrossRef] [Green Version]
- Pérez-Peña, J.; Győrffy, B.; Amir, E.; Pandiella, A.; Ocaña, A. Epigenetic modulation of FOXM1-gene interacting network by BET inhibitors in breast cancer. Breast Cancer Res. Treat. 2018, 172, 725–732. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Peña, J.; Serrano-Heras, G.; Montero, J.C.; Corrales-Sánchez, V.; Pandiella, A.; Ocaña, A. In Silico Analysis Guides Selection of BET Inhibitors for Triple-Negative Breast Cancer Treatment. Mol. Cancer Ther. 2016, 15, 1823–1833. [Google Scholar] [CrossRef] [PubMed]
- Berthon, C.; Raffoux, E.; Thomas, X.; Vey, N.; Gomez-Roca, C.; Yee, K.; Taussig, D.C.; Rezai, K.; Roumier, C.; Herait, P.; et al. Bromodomain inhibitor OTX015 in patients with acute leukaemia: A dose-escalation, phase 1 study. Lancet Haematol. 2016, 3, e186–e195. [Google Scholar] [CrossRef]
- Amorim, S.; Stathis, A.; Gleeson, M.; Iyengar, S.; Magarotto, V.; Leleu, X.; Morschhauser, F.; Karlin, L.; Broussais, F.; Rezai, K.; et al. Bromodomain inhibitor OTX015 in patients with lymphoma or multiple myeloma: A dose-escalation, open-label, pharmacokinetic, phase 1 study. Lancet Haematol. 2016, 3, e196–e204. [Google Scholar] [CrossRef]
- Kim, D.-S.; Camacho, C.V.; Kraus, W.L. Alternate therapeutic pathways for PARP inhibitors and potential mechanisms of resistance. Exp. Mol. Med. 2021, 53, 42–51. [Google Scholar] [CrossRef]
- Vikas, P.; Borcherding, N.; Chennamadhavuni, A.; Garje, R. Therapeutic Potential of Combining PARP Inhibitor and Immunotherapy in Solid Tumors. Front. Oncol. 2020, 10, 570. [Google Scholar] [CrossRef]
- Sun, C.; Fang, Y.; Yin, J.; Chen, J.; Ju, Z.; Zhang, D.; Chen, X.; Vellano, C.P.; Jeong, K.J.; Ng, P.K.-S.; et al. Rational combination therapy with PARP and MEK inhibitors capitalizes on therapeutic liabilities in RAS mutant cancers. Sci. Transl. Med. 2017, 9, eaal5148. [Google Scholar] [CrossRef]
- Yang, L.; Zhang, Y.; Shan, W.; Hu, Z.; Yuan, J.; Pi, J.; Wang, Y.; Fan, L.; Tang, Z.; Li, C.; et al. Repression of BET activity sensitizes homologous recombination-proficient cancers to PARP inhibition. Sci. Transl. Med. 2017, 9, eaal1645. [Google Scholar] [CrossRef]
- Karakashev, S.; Zhu, H.; Yokoyama, Y.; Zhao, B.; Fatkhutdinov, N.; Kossenkov, A.V.; Wilson, A.J.; Simpkins, F.; Speicher, D.; Khabele, D.; et al. BET Bromodomain Inhibition Synergizes with PARP Inhibitor in Epithelial Ovarian Cancer. Cell Rep. 2017, 21, 3398–3405. [Google Scholar] [CrossRef]
- Shorstova, T.; Foulkes, W.D.; Witcher, M. Achieving clinical success with BET inhibitors as anti-cancer agents. Br. J. Cancer 2021, 124, 1478–1490. [Google Scholar] [CrossRef]
- Maggisano, V.; Celano, M.; Malivindi, R.; Barone, I.; Cosco, D.; Mio, C.; Mignogna, C.; Panza, S.; Damante, G.; Fresta, M.; et al. Nanoparticles Loaded with the BET Inhibitor JQ1 Block the Growth of Triple Negative Breast Cancer Cells In Vitro and In Vivo. Cancers 2020, 12, 91. [Google Scholar] [CrossRef]
- Wang, Q.; Kumar, V.; Lin, F.; Sethi, B.; Coulter, D.W.; McGuire, T.R.; Mahato, R.I. ApoE mimetic peptide targeted nanoparticles carrying a BRD4 inhibitor for treating Medulloblastoma in mice. J. Control. Release 2020, 323, 463–474. [Google Scholar] [CrossRef] [PubMed]
- Xu, Q.; Wang, L.; Tong, T.; Huang, X.; Huang, C.; Li, F.; Su, Q.; Tien, Y.; Wu, J.; Zhao, W.; et al. Efficient delivery of BRD4 inhibitor by glutathione-sensitive nanoparticle to suppress gallbladder cancer through inhibiting NF-κB signaling. Appl. Mater. Today 2020, 21, 100849. [Google Scholar] [CrossRef]
- Tian, Y.; Wang, X.; Zhao, S.; Liao, X.; Younis, M.R.; Wang, S.; Zhang, C.; Lu, G. JQ1-Loaded Polydopamine Nanoplatform Inhibits c-MYC/Programmed Cell Death Ligand 1 to Enhance Photothermal Therapy for Triple-Negative Breast Cancer. ACS Appl. Mater. Interfaces 2019, 11, 46626–46636. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Chen, G.; Urabe, G.; Xie, R.; Wang, Y.; Shi, X.; Guo, L.-W.; Gong, S.; Kent, K.C. A paradigm of endothelium-protective and stent-free anti-restenotic therapy using biomimetic nanoclusters. Biomaterials 2018, 178, 293–301. [Google Scholar] [CrossRef]
- Wu, V.M.; Mickens, J.; Uskoković, V. Bisphosphonate-Functionalized Hydroxyapatite Nanoparticles for the Delivery of the Bromodomain Inhibitor JQ1 in the Treatment of Osteosarcoma. ACS Appl. Mater. Interfaces 2017, 9, 25887–25904. [Google Scholar] [CrossRef]
- Hassan, R.; Tammam, S.N.; Safy, S.E.; Abdel-Halim, M.; Asimakopoulou, A.; Weiskirchen, R.; Mansour, S. Prevention of hepatic stellate cell activation using JQ1- and atorvastatin-loaded chitosan nanoparticles as a promising approach in therapy of liver fibrosis. Eur. J. Pharm. Biopharm. 2019, 134, 96–106. [Google Scholar] [CrossRef]
- Wang, H.; Tang, Y.; Fang, Y.; Zhang, M.; Wang, H.; He, Z.; Wang, B.; Xu, Q.; Huang, Y. Reprogramming Tumor Immune Microenvironment (TIME) and Metabolism via Biomimetic Targeting Codelivery of Shikonin/JQ1. Nano Lett. 2019, 19, 2935–2944. [Google Scholar] [CrossRef]
- Zhao, H.; Song, Q.; Zheng, C.; Zhao, B.; Wu, L.; Feng, Q.; Zhang, Z.; Wang, L. Implantable Bioresponsive Nanoarray Enhances Postsurgical Immunotherapy by Activating Pyroptosis and Remodeling Tumor Microenvironment. Adv. Funct. Mater. 2020, 30, 2005747. [Google Scholar] [CrossRef]
- Zhou, F.; Gao, J.; Xu, Z.; Li, T.; Gao, A.; Sun, F.; Wang, F.; Wang, W.; Geng, Y.; Zhang, F.; et al. Overcoming immune resistance by sequential prodrug nanovesicles for promoting chemoimmunotherapy of cancer. Nano Today 2021, 36, 101025. [Google Scholar] [CrossRef]
- Lam, F.C.; Morton, S.W.; Wyckoff, J.; Vu Han, T.-L.; Hwang, M.K.; Maffa, A.; Balkanska-Sinclair, E.; Yaffe, M.B.; Floyd, S.R.; Hammond, P.T. Enhanced efficacy of combined temozolomide and bromodomain inhibitor therapy for gliomas using targeted nanoparticles. Nat. Commun. 2018, 9, 1991. [Google Scholar] [CrossRef]
- He, Z.; Zhang, M.; Wang, Y.-H.; He, Y.; Wang, H.-R.; Chen, B.-F.; Tu, B.; Zhu, S.-Q.; Huang, Y.-Z. Anti-PD-L1 mediating tumor-targeted codelivery of liposomal irinotecan/JQ1 for chemo-immunotherapy. Acta Pharm. Sin. 2021, 42, 1516–1523. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.-S.; You, X.; Dai, C.; Xu, Q.-C.; Li, F.; Wang, L.; Huang, X.-T.; Wang, J.-Q.; Li, S.-J.; Gao, Z.; et al. Targeting Super-Enhancers via Nanoparticle-Facilitated BRD4 and CDK7 Inhibitors Synergistically Suppresses Pancreatic Ductal Adenocarcinoma. Adv. Sci. 2020, 7, 1902926. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Du, C.; Zhao, Y.; Nie, G.; Yang, Y. Trap and kill strategy for non-BRCA mutant pancreatic cancer by co-delivery of olaparib and JQ1 with plectin-1 targeting peptide nanoparticles. Nano Today 2020, 33, 100877. [Google Scholar] [CrossRef]
- Broomfield, L.M.; Alonso-Moreno, C.; Martin, E.; Shafir, A.; Posadas, I.; Ceña, V.; Castro-Osma, J.A. Aminophosphine ligands as a privileged platform for development of antitumoral ruthenium(ii) arene complexes. Dalton Trans. 2017, 46, 16113–16125. [Google Scholar] [CrossRef]
- de la Cruz-Martínez, F.; Martínez de Sarasa Buchaca, M.; Martínez, J.; Tejeda, J.; Fernández-Baeza, J.; Alonso-Moreno, C.; Rodríguez, A.M.; Castro-Osma, J.A.; Lara-Sánchez, A. Bimetallic Zinc Catalysts for Ring-Opening Copolymerization Processes. Inorg. Chem. 2020, 59, 8412–8423. [Google Scholar] [CrossRef]
- Estupiñán, Ó.; Niza, E.; Bravo, I.; Rey, V.; Tornín, J.; Gallego, B.; Clemente-Casares, P.; Moris, F.; Ocaña, A.; Blanco-Lorenzo, V.; et al. Mithramycin delivery systems to develop effective therapies in sarcomas. J. Nanobiotechnol. 2021, 19, 267. [Google Scholar] [CrossRef]
- Niza, E.; Ocaña, A.; Castro-Osma, J.A.; Bravo, I.; Alonso-Moreno, C. Polyester Polymeric Nanoparticles as Platforms in the Development of Novel Nanomedicines for Cancer Treatment. Cancers 2021, 13, 3387. [Google Scholar] [CrossRef]
- Cimas, F.J.; Niza, E.; Juan, A.; Noblejas-López, M.D.; Bravo, I.; Lara-Sanchez, A.; Alonso-Moreno, C.; Ocaña, A. Controlled Delivery of BET-PROTACs: In Vitro Evaluation of MZ1-Loaded Polymeric Antibody Conjugated Nanoparticles in Breast Cancer. Pharmaceutics 2020, 12, 986. [Google Scholar] [CrossRef]
- Niza, E.; Noblejas-lópez, M.D.M.; Bravo, I.; Nieto-jiménez, C.; Castro-osma, J.A.; Canales-vázquez, J.; Lara-sanchez, A.; Moya, E.M.G.; Burgos, M.; Ocaña, A.; et al. Trastuzumab-targeted biodegradable nanoparticles for enhanced delivery of dasatinib in HER2+ metastasic breast cancer. Nanomaterials 2019, 9, 1793. [Google Scholar] [CrossRef] [Green Version]
- Cabral, H.; Kataoka, K. Progress of drug-loaded polymeric micelles into clinical studies. J. Control. Release 2014, 190, 465–476. [Google Scholar] [CrossRef] [PubMed]
- Nuncia Cantarero, M.; Serrano-Oviedo, L.; Nieto Jiménez, C.; Tébar García, D.; Burgos Lozano, M.; Morcillo García, S.; Perez Peña, J.; Corrales-Sanchez, V.; Gyorffy, B.; Ocana, A.; et al. Identification of a stemness-related gene panel associated with BET inhibition in triple negative breast cancer. JCO 2020, 38, e13098. [Google Scholar] [CrossRef]
- Amir, E.; Seruga, B.; Serrano, R.; Ocana, A. Targeting DNA repair in breast cancer: A clinical and translational update. Cancer Treat. Rev. 2010, 36, 557–565. [Google Scholar] [CrossRef] [PubMed]
- Morel, D.; Jeffery, D.; Aspeslagh, S.; Almouzni, G.; Postel-Vinay, S. Combining epigenetic drugs with other therapies for solid tumours—Past lessons and future promise. Nat. Rev. Clin. Oncol. 2020, 17, 91–107. [Google Scholar] [CrossRef]
- Ocaña, A.; Nieto-Jiménez, C.; Pandiella, A. BET inhibitors as novel therapeutic agents in breast cancer. Oncotarget 2017, 8, 71285–71291. [Google Scholar] [CrossRef]
- Thi, T.T.H.; Suys, E.J.A.; Lee, J.S.; Nguyen, D.H.; Park, K.D.; Truong, N.P. Lipid-Based Nanoparticles in the Clinic and Clinical Trials: From Cancer Nanomedicine to COVID-19 Vaccines. Vaccines 2021, 9, 359. [Google Scholar] [CrossRef]
- Pramanik, A.; Xu, Z.; Shamsuddin, S.H.; Khaled, Y.S.; Ingram, N.; Maisey, T.; Tomlinson, D.; Coletta, P.L.; Jayne, D.; Hughes, T.A.; et al. Affimer Tagged Cubosomes: Targeting of Carcinoembryonic Antigen Expressing Colorectal Cancer Cells Using In Vitro and In Vivo Models. ACS Appl. Mater. Interfaces 2022, 14, 11078–11091. [Google Scholar] [CrossRef]
- Pramanik, A.; Xu, Z.; Ingram, N.; Coletta, P.L.; Millner, P.A.; Tyler, A.I.I.; Hughes, T.A. Hyaluronic-Acid-Tagged Cubosomes Deliver Cytotoxics Specifically to CD44-Positive Cancer Cells. Mol. Pharm. 2022. [Google Scholar] [CrossRef]
- Hutchinson, S.A.; Websdale, A.; Cioccoloni, G.; Røberg-Larsen, H.; Lianto, P.; Kim, B.; Rose, A.; Soteriou, C.; Pramanik, A.; Wastall, L.M.; et al. Liver x receptor alpha drives chemoresistance in response to side-chain hydroxycholesterols in triple negative breast cancer. Oncogene 2021, 40, 2872–2883. [Google Scholar] [CrossRef]
- Baxter, D.E.; Allinson, L.M.; Al Amri, W.S.; Poulter, J.A.; Pramanik, A.; Thorne, J.L.; Verghese, E.T.; Hughes, T.A. MiR-195 and Its Target SEMA6D Regulate Chemoresponse in Breast Cancer. Cancers 2021, 13, 5979. [Google Scholar] [CrossRef]








| Formulation | RH (nm) | PDI | Z Potential (mV) |
|---|---|---|---|
| JQ1-LP | 61.04 ± 0.14 | 0.13 ± 0.02 | −34.20 ± 1.75 |
| JQ1-NPs | 125.19 ± 1.67 | 0.08 ± 0.01 | −15.93 ± 0.60 |
| NPs | 138.63 ± 9.29 | 0.09 ± 0.02 | 0.94 ± 0.27 |
| LIP | 65.21 ± 1.08 | 0.08 ± 0.02 | −35.00 ± 2.27 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Juan, A.; Noblejas-López, M.d.M.; Bravo, I.; Arenas-Moreira, M.; Blasco-Navarro, C.; Clemente-Casares, P.; Lara-Sánchez, A.; Pandiella, A.; Alonso-Moreno, C.; Ocaña, A. Enhanced Antitumoral Activity of Encapsulated BET Inhibitors When Combined with PARP Inhibitors for the Treatment of Triple-Negative Breast and Ovarian Cancers. Cancers 2022, 14, 4474. https://doi.org/10.3390/cancers14184474
Juan A, Noblejas-López MdM, Bravo I, Arenas-Moreira M, Blasco-Navarro C, Clemente-Casares P, Lara-Sánchez A, Pandiella A, Alonso-Moreno C, Ocaña A. Enhanced Antitumoral Activity of Encapsulated BET Inhibitors When Combined with PARP Inhibitors for the Treatment of Triple-Negative Breast and Ovarian Cancers. Cancers. 2022; 14(18):4474. https://doi.org/10.3390/cancers14184474
Chicago/Turabian StyleJuan, Alberto, María del Mar Noblejas-López, Iván Bravo, María Arenas-Moreira, Cristina Blasco-Navarro, Pilar Clemente-Casares, Agustín Lara-Sánchez, Atanasio Pandiella, Carlos Alonso-Moreno, and Alberto Ocaña. 2022. "Enhanced Antitumoral Activity of Encapsulated BET Inhibitors When Combined with PARP Inhibitors for the Treatment of Triple-Negative Breast and Ovarian Cancers" Cancers 14, no. 18: 4474. https://doi.org/10.3390/cancers14184474
APA StyleJuan, A., Noblejas-López, M. d. M., Bravo, I., Arenas-Moreira, M., Blasco-Navarro, C., Clemente-Casares, P., Lara-Sánchez, A., Pandiella, A., Alonso-Moreno, C., & Ocaña, A. (2022). Enhanced Antitumoral Activity of Encapsulated BET Inhibitors When Combined with PARP Inhibitors for the Treatment of Triple-Negative Breast and Ovarian Cancers. Cancers, 14(18), 4474. https://doi.org/10.3390/cancers14184474