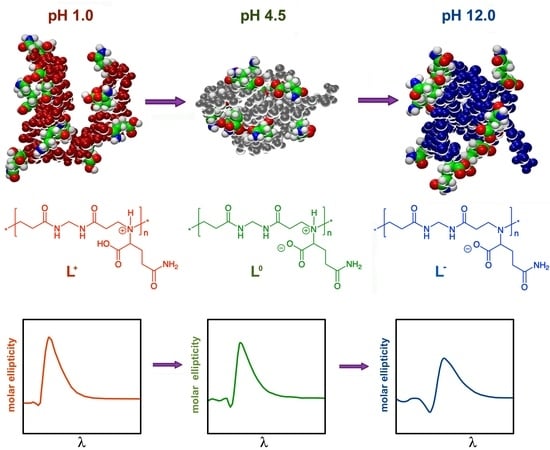
Hydrogen Bonding in a l-Glutamine-Based Polyamidoamino Acid and its pH-Dependent Self-Ordered Coil Conformation
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Characterizations
2.3. Synthesis of M-l-Gln
2.4. Molecular Mechanics (MM) and Molecular Dynamics (MD) Simulations
3. Results and Discussion
3.1. Synthesis of M-l-Gln
3.2. Acid–Base Properties of M-l-Gln
3.3. Dynamic Light Scattering (DLS) Measurements
3.4. Circular Dichroism Spectrocopy
3.5. Molecular Simulation Studies
3.5.1. Molecular Mechanics: Conformations of the Optimized Geometries
3.5.2. Molecular Dynamics: H-Bond Distribution at pH 4.5
3.6. NMR Analyses
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Philp, D.; Stoddart, J.F. Self-assembly in natural and unnatural systems. Angew. Chem. Int. Ed. 1996, 35, 1154–1196. [Google Scholar] [CrossRef]
- Mendes, A.C.; Baran, E.T.; Reis, R.L.; Azevedo, H.S. Self-assembly in nature: Using the principles of nature to create complex nanobiomaterials. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 2013, 5, 582–612. [Google Scholar] [CrossRef] [PubMed]
- Whitesides, G.M.; Grzybowski, B. Self-assembly at all scales. Science 2002, 295, 2418–2421. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garcia, M.A.; Iglesias, D.; Parisi, E.; Styan, K.E.; Waddington, L.J.; Deganutti, C.; De Zorzi, R.; Grassi, M.; Melchionna, M.; Vargiu, A.V.; et al. Chirality effects on peptide self-assembly unraveled from molecules to materials. CHEM 2018, 4, 1862–1876. [Google Scholar] [CrossRef] [Green Version]
- Marchesan, S.; Waddington, L.; Easton, C.D.; Winkler, D.A.; Goodall, L.; Forsythe, J.; Hartley, P.G. Unzipping the role of chirality in nanoscale self-assembly of tripeptide hydrogels. Nanoscale 2012, 4, 6752–6760. [Google Scholar] [CrossRef]
- Marchesan, S.; Easton, C.D.; Kushkaki, F.; Waddington, L.; Hartley, P.G. Tripeptide self-assembled hydrogels: Unexpected twists of chirality. Chem. Commun. 2012, 48, 2195–2197. [Google Scholar] [CrossRef]
- Burke, M.G.; Woscholski, R.; Yaliraki, S.N. Differential hydrophobicity drives self-assembly in Huntington’s disease. Proc. Natl. Acad. Sci. USA 2003, 100, 13928–13933. [Google Scholar] [CrossRef] [Green Version]
- Pan, S.; Liu, W.; Tang, J.; Yang, Y.; Feng, H.; Qian, Z.; Zhou, J. Hydrophobicity-guided self-assembled particles of silver nanoclusters with aggregation-induced emission and their use in sensing and bioimaging. J. Mater. Chem. B 2018, 6, 3927–3933. [Google Scholar] [CrossRef]
- Yang, P.; Zhao, X.; Xu, A.; Wang, W.; Wang, H. Reorganization of self-assembled supramolecular materials controlled by hydrogen bonding and hydrophilic–lipophilic balance. J. Mater. Chem. B 2016, 4, 2662–2668. [Google Scholar] [CrossRef]
- Kuo, S.W. Hydrogen bond-mediated self-assembly and supramolecular structures of diblock copolymer mixtures. Polym. Int. 2009, 58, 455–464. [Google Scholar] [CrossRef]
- Lefevre, N.; Fustin, L.A.; Gohy, J.F. Reorganization of hydrogen-bonded block copolymer complexes. Langmuir 2007, 23, 4618–4622. [Google Scholar] [CrossRef] [PubMed]
- Bauri, K.; Ghosh Roy, S.; De, P. Side chain amino-acid-derived cationic chiral polymers by controlled radical polymerization. Macromol. Chem. Phys. 2016, 217, 365–379. [Google Scholar] [CrossRef]
- Wang, X.; Gan, H.; Sun, T.; Su, B.; Fuchs, H.; Vestweber, D.; Butz, S. Stereochemistry triggered differential cell behaviors on chiral polymer surfaces. Soft Matter 2010, 6, 3851–3855. [Google Scholar] [CrossRef]
- Sinaga, A.; Hatton, T.A.; Tam, K.C. Poly(acrylic acid)block-poly(L-valine): Evaluation of β-sheet formation and its stability using circular dichroism technique. Biomacromolecules 2007, 8, 2801–2808. [Google Scholar] [CrossRef]
- Bloksma, M.M.; Rogers, S.; Schubert, U.S.; Hoogenboom, R. Secondary structure formation of main chain chiral poly(2-oxazoline)s in solution. Soft Matter 2010, 6, 994–1003. [Google Scholar] [CrossRef]
- Luo, R.; Zhu, M.; Shen, X.; Li, S. Polymer catalyst with self-assembled hierarchical access for sortable catalysis. J. Catal. 2015, 331, 49–56. [Google Scholar] [CrossRef]
- Evans, C.A.; Skey, J.; Wright, M.; Qu, W.; Ondeck, C.; Longbottom, D.A.; O’ Reilly, R.K. Functional and tuneable amino acid polymers prepared by RAFT polymerization. J. Polym. Sci. A 2009, 47, 6814–6826. [Google Scholar] [CrossRef]
- Kristensen, T.E.; Vestli, K.; Jakobsen, M.G.; Hansen, F.K.; Hansen, T. A general approach for preparation of polymer-supported chiral organocatalysts via acrylic copolymerization. J. Org. Chem. 2010, 75, 1620–1629. [Google Scholar] [CrossRef] [PubMed]
- Itsuno, S.; Hassan, M. Polymer-immobilized chiral catalysts. RSC Adv. 2014, 4, 52023–52043. [Google Scholar] [CrossRef]
- Liang, J.; Yang, B.; Deng, J. Polylactide-based chiral particles with enantio-differentiating release ability. Chem. Eng. Sci. 2018, 344, 262–269. [Google Scholar] [CrossRef]
- Quiñones, J.P.; Peniche, H.; Peniche, C. Chitosan based self-assembled nanoparticles in drug delivery. Polymers 2018, 10, 235. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morioka, K.; Suito, Y.; Isobe, Y.; Habaue, S.; Okamoto, Y. Synthesis and chiral recognition ability of optically active poly{N-[(R)-α-methoxycarbonylbenzyl]methacrylamide} with various tacticities by radical polymerization using Lewis acids. J. Polym. Sci. A Polym. Chem. 2003, 41, 3354–3360. [Google Scholar] [CrossRef]
- Shen, J.; Okamoto, Y. Efficient separation of enantiomers using stereoregular chiral polymers. Chem. Rev. 2016, 116, 1094–1138. [Google Scholar] [CrossRef] [PubMed]
- Mastai, Y.; Sedlak, M.; Colfen, H.; Antonietti, M. The separation of racemic crystals into enantiomers by chiral block copolymers. Chem. Eur. J. 2002, 8, 2430–2437. [Google Scholar] [CrossRef]
- Menahem, T.; Mastai, Y. Chiral soluble polymers and microspheres for enantioselective crystallization. J. Polym. Sci. A Polym. Chem. 2006, 44, 3009–3017. [Google Scholar] [CrossRef]
- Menahem, T.; Pravda, M.; Mastai, Y. Correlation between structures of chiral polymers and their efficiency for chiral resolution by crystallization. Chirality 2009, 21, 862–870. [Google Scholar] [CrossRef]
- Fukuhara, G. Polymer-based supramolecular sensing and application to chiral photochemistry. Polym. J. 2015, 47, 649–655. [Google Scholar] [CrossRef]
- Fukuhara, G.; Inoue, Y. Chirality-sensing binaphthocrown ether–polythiophene conjugate. Chem. Eur. J. 2010, 16, 7859–7864. [Google Scholar] [CrossRef]
- Yashima, E.; Maeda, K.; Nishimura, T. Detection and amplification of chirality by helical polymers. Chem. Eur. J. 2004, 10, 42–51. [Google Scholar] [CrossRef]
- Dai, C.; Yang, D.; Zhang, W.; Bao, B.; Cheng, Y.; Wang, L. Far-red/near-infrared fluorescent conjugated polymer nanoparticles with size-dependent chirality and cell imaging applications. Polym. Chem. 2015, 6, 3962–3969. [Google Scholar] [CrossRef]
- Ferruti, P.; Mauro, N.; Falciola, L.; Pifferi, V.; Bartoli, C.; Gazzarri, M.; Chiellini, F.; Ranucci, E. Amphoteric, prevailingly cationic L-arginine polymers of poly(amidoamino acid) structure: Synthesis, acid/base properties and preliminary cytocompatibility and cell-permeating characterizations. Macromol. Biosci. 2014, 14, 390–400. [Google Scholar] [CrossRef] [PubMed]
- Manfredi, A.; Mauro, N.; Terenzi, A.; Alongi, J.; Lazzari, F.; Ganazzoli, F.; Raffaini, G.; Ranucci, E.; Ferruti, P. Self-ordering secondary structure of d- and l-arginine-derived polyamidoamino acids. ACS Macro Lett. 2017, 6, 987–991. [Google Scholar] [CrossRef]
- Lazzari, F.; Manfredi, A.; Alongi, J.; Mendichi, R.; Ganazzoli, F.; Raffaini, G.; Ferruti, P.; Ranucci, E. Self-structuring in water of polyamidoamino acids with hydrophobic side chains deriving from natural α-amino acids. Polymers 2018, 10, 1261. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lazzari, F.; Manfredi, A.; Alongi, J.; Marinotto, D.; Ferruti, P.; Ranucci, E. d-, l- and d,l-tryptophan-based polyamidoamino Acids: pH-dependent structuring and fluorescent properties. Polymers 2019, 11, 543. [Google Scholar] [CrossRef] [Green Version]
- Rhys, N.H.; Soper, A.K.; Dougan, L. The hydrogen-bonding ability of the amino acid glutamine revealed by neutron diffraction scattering experiments. J. Phys. Chem. B 2012, 116, 13308–13319. [Google Scholar] [CrossRef]
- Rhys, N.H.; Dougan, L. The emerging role of hydrogen bond interactions in polyglutamine structure, stability and association. Soft Matter 2013, 9, 2359–2364. [Google Scholar] [CrossRef]
- Mele, A.; Ganazzoli, F.; Raffaini, G.; Juza, M.; Schurig, V. Macrocycle conformation and self-inclusion phenomena in octakis(3-O-butanoyl-2,6-di-O-n-pentyl)-γ-cyclodextrin (Lipodex E) by NMR spectroscopy and molecular dynamics. Carbohydr. Res. 2003, 338, 625–635. [Google Scholar] [CrossRef]
- Raffaini, G.; Ganazzoli, F. Hydration and flexibility of α-, β-, γ- and δ-cyclodextrin: A molecular dynamics study. Chem. Phys. 2003, 333, 625–635. [Google Scholar] [CrossRef]
- Raffaini, G.; Ganazzoli, F.; Malpezzi, L.; Fuganti, C.; Fronza, G.; Panzeri, W.; Mele, A.A. Molecular Dynamics Study of the Inclusion Complexes of C60 with Some Cyclodextrins. J. Phys. Chem. B 2009, 113, 9110–9122. [Google Scholar] [CrossRef]
- El Bouakher, A.; Martel, A.; Comesse, S. α-Halogenoacetamides: Versatile and efficient tools for the synthesis of complex aza-heterocycles. Org. Biomol. Chem. 2019, 17, 8467–8485. [Google Scholar] [CrossRef]
- Lee, A.S.; Lin, M.-C.; Lin, C.-C.; Chang, Y.-T. Me3SiOTf Promoted Aza-Michael Addition Reaction of 1°-Amides with α,β-Unsaturated Ketones. J. Chin. Chem. Soc. 2010, 57, 795–799. [Google Scholar] [CrossRef]
- Katchalsky, A.; Spitnik, P. Potentiometric titrations of polymethacrylic acid. J. Polym. Sci. 1947, 2, 432–446. [Google Scholar] [CrossRef]
- Kelly, S.M.; Price, N.C. The use of circular dichroism in the investigation of protein structure and function. Curr. Protein Pept. Sci. 2000, 1, 349–384. [Google Scholar] [CrossRef] [Green Version]
- Evans, R.; Deng, Z.; Rogerson, A.K.; McLachlan, A.S.; Richards, J.J.; Nilsson, M.; Morris, G.A. Quantitative Interpretation of Diffusion-Ordered NMR Spectra: Can We Rationalize Small Molecule Diffusion Coefficients? Angew. Chem. 2013, 125, 3281–3284. [Google Scholar] [CrossRef]
- Marchini, M.; Mingozzi, M.; Colombo, R.; Guzzetti, I.; Belvisi, L.; Vasile, F.; Potenza, D.; Piarulli, U.; Arosio, D.; Gennari, C. Cyclic RGD peptidomimetics containing bifunctional diketopiperazine scaffolds as new potent integrin ligands. Chem. A Eur. J. 2012, 18, 6195–6207. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vasile, F.; Rossi, D.; Collina, S.; Potenza, D. Diffusion-Ordered Spectroscopy and Saturation Transfer Difference NMR Spectroscopy Studies of Selective Interactions between ELAV Protein Fragments and a mRNA Target. Eur. J. Org. Chem. 2014, 29, 6399–6404. [Google Scholar] [CrossRef]
- Vasile, F.; Civera, M.; Belvisi, L.; Potenza, D.; Tiana, G. Thermodynamically–weighted conformational ensemble of cyclic RGD peptidomimetics from NOE data. J. Phys. Chem. B 2016, 120, 7098–7107. [Google Scholar] [CrossRef] [PubMed]
- Bovey, F.A.; Mirau, P.A. The Solution Characterization of Polymers. In NMR of Polymers; Academic Press: Cambridge, MA, USA, 1999; pp. 155–241. [Google Scholar] [CrossRef]
- Gellman, S.H.; Dado, G.P.; Liang, G.B.; Adams, B.R. Conformation-directing effects of a single intramolecular amide-amide hydrogen bond: Variable-temperature NMR and IR studies on a homologous diamide series. J. Am. Chem. Soc. 1991, 113, 1164–1173. [Google Scholar] [CrossRef]











| Sample | Mwb | PDI c |
|---|---|---|
| M-l-Gln | 21500 | 2.31 |
| M-l-Ala d | 7800 | 1.44 |
| M-l-Val d | 5600 | 1.30 |
| M-l-Leu d | 6200 | 1.51 |
| pKa1b | pKa2c | β1b | β2c | IP d |
|---|---|---|---|---|
| 2.17 ± 0.05 | 6.80 ± 0.05 | 0.84 ± 0.09 | 1.62 ± 0.08 | 4.5 |
| pH | Charge | Rga (nm) | Sb (nm2) | Volume c (nm3) | μd (D) |
|---|---|---|---|---|---|
| 1.0 | L+ | 1.07 | 25.1 | 2.91 | 19.8 |
| 4.5 | L0 | 0.82 | 17.1 | 3.23 | 32.1 |
| 12.0 | L− | 0.98 | 23.5 | 2.96 | 23.2 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lazzari, F.; Manfredi, A.; Alongi, J.; Ganazzoli, F.; Vasile, F.; Raffaini, G.; Ferruti, P.; Ranucci, E. Hydrogen Bonding in a l-Glutamine-Based Polyamidoamino Acid and its pH-Dependent Self-Ordered Coil Conformation. Polymers 2020, 12, 881. https://doi.org/10.3390/polym12040881
Lazzari F, Manfredi A, Alongi J, Ganazzoli F, Vasile F, Raffaini G, Ferruti P, Ranucci E. Hydrogen Bonding in a l-Glutamine-Based Polyamidoamino Acid and its pH-Dependent Self-Ordered Coil Conformation. Polymers. 2020; 12(4):881. https://doi.org/10.3390/polym12040881
Chicago/Turabian StyleLazzari, Federica, Amedea Manfredi, Jenny Alongi, Fabio Ganazzoli, Francesca Vasile, Giuseppina Raffaini, Paolo Ferruti, and Elisabetta Ranucci. 2020. "Hydrogen Bonding in a l-Glutamine-Based Polyamidoamino Acid and its pH-Dependent Self-Ordered Coil Conformation" Polymers 12, no. 4: 881. https://doi.org/10.3390/polym12040881
APA StyleLazzari, F., Manfredi, A., Alongi, J., Ganazzoli, F., Vasile, F., Raffaini, G., Ferruti, P., & Ranucci, E. (2020). Hydrogen Bonding in a l-Glutamine-Based Polyamidoamino Acid and its pH-Dependent Self-Ordered Coil Conformation. Polymers, 12(4), 881. https://doi.org/10.3390/polym12040881