Comprehensive Leaf Cell Wall Analysis Using Carbohydrate Microarrays Reveals Polysaccharide-Level Variation between Vitis Species with Differing Resistance to Downy Mildew
Abstract
:1. Introduction
2. Material and Methods
2.1. Plant Materials and Disease Resistance Assessment
2.2. Cell Wall Preparation
2.3. Comprehensive Microarray Polymer Profiling (CoMPP)
2.4. Gas Chromatography–Mass Spectrometry (GC–MS) for Monosaccharides
2.5. Fourier Transform Infrared (FTIR) Spectroscopy
2.6. Univariate Statistical and Multivariate Data Analysis
3. Results and Discussion
3.1. Downy Mildew Resistance Measurement of Leaf Disks Sourced from the Two Grapevine Cultivars and the Hybrid Cross
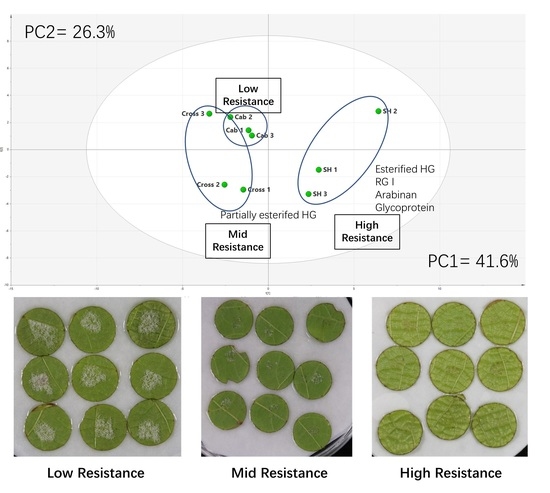
3.2. Comprehensive Microarray Polymer Profiling (CoMPP)
3.3. Monosaccharide Composition of Leaf Cell Wall Materials Using GC–MS
3.4. IR Spectroscopy
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Moore, J.P.; Divol, B. Tracking the careers of grape and wine polymers using biotechnology and systems biology. In Biotechnology in Functional Foods and Nutraceuticals; Bagchi, D., Lau, F.C., Ghosh, D.K., Eds.; CRC Press Inc.: Boca Raton, FL, USA, 2010; pp. 389–406. [Google Scholar]
- Kubicek, C.P.; Starr, T.L.; Glass, N.L. Plant cell wall-degrading enzymes and their secretion in plant-pathogen fungi. Annu. Rev. Phytopathol. 2014, 52, 427–451. [Google Scholar] [CrossRef] [PubMed]
- Boevink, P.C.; Mclellan, H.; Gilroy, E.M.; Naqvi, S.; He, Q.; Yang, L.; Wang, X.; Turnbull, D.; Armstrong, M.R.; Tian, Z.; et al. Oomycetes seek help from the plant: Phytophthora infestans effectors target host susceptibility factors. Mol. Plant 2016, 9, 636–638. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Johnsen, H.R.; Striberny, B.; Olsen, S.; Vidal-Melgosa, S.; Fangel, J.U.; Willats, W.G.T.; Rose, J.K.C.; Krause, K. Cell wall composition profiling of parasitic giant dodder (Cuscuta reflexa) and its hosts: A priori differences and induced changes. New Phytol. 2015, 207, 805–816. [Google Scholar] [CrossRef]
- Olsen, S.; Striberny, B.; Hollmann, J.; Schwacke, R.; Popper, Z.A.; Krause, K. Getting ready for host invasion elevated expression and action of xyloglucan endotransglucosylases/hydrolases in developing haustoria of the holoparasitic angiosperm Cuscuta. J. Exp. Bot. 2016, 67, 695–708. [Google Scholar] [CrossRef] [Green Version]
- Bacete, L.; Melida, H.; Miedes, E.; Molina, A. Plant cell wall-mediated immunity: Cell wall changes trigger disease resistance responses. Plant J. 2018, 93, 614–636. [Google Scholar] [CrossRef]
- Beck, M.; Zhou, J.; Faulkner, C.; Maclean, D.; Robatzek, S. Spatio-temporal cellular dynamics of the Arabidopsis flagellin receptor reveal activation status-dependent endosomal sorting. Plant Cell. 2012, 24, 4205. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Jonge, R.; van Esse, H.P.; Kombrink, A.; Shinya, T.; Desaki, Y.; Bours, R.; Van Der Krol, S.; Shibuya, N.; Joosten, M.H.A.J.; Thomma, B.P.H.J. Conserved fungal lysm effector ecp6 prevents chitin-triggered immunity in plants. Science 2010, 329, 953. [Google Scholar] [CrossRef]
- Hernández Blanco, C.; Feng, D.X.; Hu, J.; Sánchez vallet, A.; Deslandes, L.; Llorente, F.; Berrocal-Lobo, M.; Keller, H.; Barlet, X.; Sánchez-Rodríguez, C.; et al. Impairment of cellulose synthases required for Arabidopsis secondary cell wall formation enhances disease resistance. Plant Cell. 2007, 19, 890–903. [Google Scholar] [CrossRef] [Green Version]
- Sicilia, F.; Fernandez-Recio, J.; Caprari, C.; De Lorenzo, G.; Tsernoglou, D.; Cervone, F.; Federici, L. The Polygalacturonase-inhibiting protein pgip2 of Phaseolus vulgaris has evolved a mixed mode of inhibition of endopolygalacturonase pg1 of Botrytis cinerea 1. Plant Physiol. 2005, 139, 1380–1388. [Google Scholar] [CrossRef] [Green Version]
- Joubert, D.A.; Kars, I.; Wagemakers, L.; Bergmann, C.; Kemp, G.; Vivier, M.A.; Van Kan, J.A.L. A polygalacturonase-inhibiting protein from grapevine reduces the symptoms of the endopolygalacturonase BcPG2 from Botrytis cinerea in Nicotiana benthamiana leaves without any evidence for in vitro interaction. Mol. Plant-Microbe Interact. 2007, 20, 392–402. [Google Scholar] [CrossRef] [Green Version]
- Federici, L.; Caprari, C.; Mattei, B.; Savino, C.; Di Matteo, A.; De Lorenzo, G.; Cervone, F.; Tsernoglou, D.; Staskawicz, B.J. Structural requirements of endopolygalacturonase for the interaction with PGIP (polygalacturonase- inhibiting protein). Proc. Natl. Acad. Sci. USA 2001, 98, 13425–13430. [Google Scholar] [CrossRef] [Green Version]
- Alexandersson, E.; Becker, J.V.W.; Jacobson, D.; Nguema-Ona, E.; Steyn, C.; Denby, K.J.; Vivier, M.A. Constitutive expression of a grapevine polygalacturonase-inhibiting protein affects gene expression and cell wall properties in uninfected tobacco. BMC Res. Notes. 2011, 4, 493. [Google Scholar] [CrossRef] [Green Version]
- Liu, N.; Zhang, X.; Sun, Y.; Wang, P.; Li, X.; Pei, Y.; Li, F.; Hou, Y. Molecular evidence for the involvement of a polygalacturonase-inhibiting protein, GhPGIP1, in enhanced resistance to Verticillium and Fusarium wilts in cotton. Sci. Rep. 2017, 7, 39840. [Google Scholar] [CrossRef] [Green Version]
- Yu, Y.; Zhang, Y.L.; Yin, L.; Lu, J. The mode of host resistance to Plasmopara viticola infection of grapevines. Phytopathol. 2012, 102, 1094–1101. [Google Scholar] [CrossRef] [Green Version]
- Burruano, S. The life-cycle of Plasmopara viticola, cause of downy mildew of vine. Mycologist 2000, 14, 179–182. [Google Scholar] [CrossRef]
- Boso, S.; Santiago, J.L.; Martinez, M.C. Resistance of eight different clones of the grape cultivar Albariño to Plasmopara viticola. Plant Dis. 2004, 88, 741–744. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Davidson, L.C. Variation within and between Vitis spp. for foliar resistance to the downy mildew pathogen Plasmopara viticola. Plant Dis. 2008, 92, 1577–1584. [Google Scholar] [CrossRef] [Green Version]
- Unger, S.; Büche, C.; Boso, S.; Kassemeyer, H.H. The course of colonization of two different Vitis genotypes by Plasmopara viticola indicates compatible and incompatible host–pathogen interactions. Phytopathology 2007, 97, 780–786. [Google Scholar] [CrossRef] [Green Version]
- Trouvelot, S.; Varnier, A.L.; Allègre, M.; Mercier, L.; Baillieul, F.; Arnould, C.; Gianinazzi-Pearson, V.; Klarzynski, O.; Joubert, J.M.; Pugin, A. A β-1,3 glucan sulfate induces resistance in grapevine against Plasmopara viticola through priming of defense responses, including HR-like cell death. Mol. Plant-Microbe Interact. 2008, 21, 232–243. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, J.; Zhang, Y.L.; Zhang, H.Q.; Huang, H.; Folta, K.M.; Lu, J. Whole genome wide expression profiles of Vitis amurensis grape responding to downy mildew by using Solexa sequencing technology. BMC Plant Biol. 2010, 10, 234–249. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vorwerk, S.; Somerville, S.; Somerville, C. The role of plant cell wall polysaccharide composition in disease resistance. Trends Plant Sci. 2004, 9, 203–209. [Google Scholar] [CrossRef]
- Somerville, C.; Bauer, S.; Brininstool, G.; Facette, M.; Hamann, T.; Milne, J.; Osborne, E.; Paredez, A.; Persson, S.; Raab, T.; et al. Toward a systems approach to understanding plant cell walls. Science 2004, 306, 2206–2211. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Albersheim, P.; Darvill, A.; Roberts, K.; Sederoff, R.; Staehelin, A. Plant Cell Walls: From Chemistry to Biology; Garland Science: New York, NY, USA, 2011; pp. 365–407. [Google Scholar]
- Marcus, S.E.; Blake, A.W.; Benians, T.A.S.; Lee, K.J.D.; Poyser, C.; Donaldson, L.; Leroux, O.; Rogowski, A.; Petersen, H.L.; Boraston, A.; et al. Restricted access of proteins to mannan polysaccharides in intact plant cell walls. Plant J. 2010, 64, 191–203. [Google Scholar] [CrossRef]
- Ma, Q.-H.; Zhu, H.-H.; Qiao, M.-Y. Contribution of both lignin content and sinapyl monomer to disease resistance in tobacco. Plant Pathol. 2018, 67, 642–650. [Google Scholar] [CrossRef]
- Yang, Y.; Yang, X.; Dong, Y.; Qiu, D. The Botrytis cinerea Xylanase BcXyl1 Modulates Plant Immunity. Front. Microbiol. 2018, 9, 2535. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moller, I.; Sørensen, I.; Bernal, A.; Blaukopf, C.; Lee, K.; Øbro, J.; Pettolino, F.; Roberts, A.; Mikkelsen, J.D.; Knox, J.P.; et al. Highthroughput mapping of cell-wall polymers within and between plants using novel microarrays. Plant J. 2007, 50, 1118–1128. [Google Scholar] [CrossRef]
- Gao, Y.; Fangel, J.U.; Willats, W.G.T.; Vivier, M.A.; Moore, J.P. Effect of commercial enzymes on berry cell wall deconstruction in the context of intravineyard ripeness variation under winemaking conditions. J. Agric. Food Chem. 2016, 64, 3862–3872. [Google Scholar] [CrossRef]
- Moore, J.P.; Fangel, J.U.; Willats, W.G.T.; Vivier, M.A. Pectic-(1,4)-galactan, extensin and arabinogalactan-protein epitopes differentiate ripening stages in wine and table grape cell walls. Ann. Bot. 2014, 114, 1279–1294. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, Y.; Fangel, J.U.; Willats, W.G.T.; Vivier, M.A.; Moore, J.P. Dissecting the polysaccharide-rich grape cell wall changes during winemaking using combined high-throughput and fractionation methods. Carbohydr. Polym. 2015, 133, 567–577. [Google Scholar] [CrossRef] [PubMed]
- Zietsman, A.J.J.; Moore, J.P.; Fangel, J.U.; Willats, W.G.T.; Vivier, M.A. Profiling the hydrolysis of isolated grape berry skin cell walls by purified enzymes. J. Agric. Food Chem. 2015, 63, 8267–8274. [Google Scholar] [CrossRef] [PubMed]
- Moore, J.P.; Nguema-Ona, E.; Fangel, J.U.; Willats, W.G.; Hugo, A.; Vivier, M.A. Profiling the main cell wall polysaccharides of grapevine leaves using high-throughput and fractionation techniques. Carbohydr. Polym. 2014, 99, 190–198. [Google Scholar] [CrossRef]
- Garrido-Bañuelos, G.; Buica, A.; Schückel, J.; Zietsman, A.J.J.; Willats, W.G.T.; Moore, J.P.; Du Toit, W.J. Investigating the relationship between grape cell wall polysaccharide composition and the extractability of phenolic compounds into Shiraz wines. Part I: Vintage and ripeness effects. Food Chem. 2019, 278, 36–46. [Google Scholar] [CrossRef]
- Ralet, M.C.; Tranquet, O.; Poulain, D.; Moise, A.; Guillon, F. Monoclonal antibodies to rhamnogalacturonan I backbone. Planta 2010, 231, 1373–1383. [Google Scholar] [CrossRef]
- Nguema-Ona, E.; Moore, J.P.; Fagerstrom, A.; Fangel, J.U.; Willats, W.G.; Hugo, A.; Vivier, M.A. Profiling the main cell wall polysaccharides of tobacco leaves using high-throughput and fractionation techniques. Carbohydr. Polym. 2012, 88, 939–949. [Google Scholar] [CrossRef]
- Gao, Y.; Fangel, J.U.; Willats, W.G.T.; Vivier, M.A.; Moore, J.P. Dissecting the polysaccharide-rich grape cell wall matrix using recombinant pectinases during winemaking. Carbohydr. Polym. 2016, 152, 510–519. [Google Scholar] [CrossRef]
- Marcus, S.E.; Verhertbruggen, Y.; Herve, C.; Ordaz-Ortiz, J.J.; Farkas, V.; Pedersen, H.L.; Willats, W.G.T.; Knox, J.P. Pectic homogalacturonan masks abundant sets of xyloglucan epitopes in plant cell walls. BMC Plant Biol. 2008, 8, 60. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Herve, C.; Rogowski, A.; Blake, A.W.; Marcus, S.E.; Gilbert, H.J.; Knox, J.P. Carbohydrate-binding modules promote the enzymatic deconstruction of intact plant cell walls by targeting and proximity effects. Proc. Natl. Acad. Sci. USA 2010, 107, 15293–15298. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tingley, J.P.; Low, K.E.; Xing, X.; Abbott, D.W. Combined whole cell wall analysis and streamlined in silico carbohydrate-active enzyme discovery to improve biocatalytic conversion of agricultural crop residues. Biotechnol. Biofuels 2021, 14, 16. [Google Scholar] [CrossRef]
- Volpi, C.; Janni, M.; Lionetti, V.; Bellincampi, D.; Favaron, F.; D’Ovidio, R. The ectopic expression of a pectin methyl esterase inhibitor increases pectin methyl esterification and limits fungal diseases in wheat. Mol. Plant-Microbe Interact. 2011, 24, 1012–1019. [Google Scholar] [CrossRef] [Green Version]
- Liu, N.; Sun, Y.; Pei, Y.; Zhang, X.; Wang, P.; Li, X.; Li, F.; Hou, Y. A pectin methylesterase inhibitor enhances resistance to verticillium wilt. Plant Physiol. 2018, 176, 2202–2220. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Coenen, G.J.; Bakx, E.J.; Verhoef, R.P.; Schols, H.A.; Voragen, A.G.J. Identification of the connecting linkage between homo- or xylogalacturonan and rhamnogalacturonan type I. Carbohydr. Polym. 2007, 70, 224–235. [Google Scholar] [CrossRef]
- Zykwinska, A.; Thibault, J.F.; Ralet, M.C. Organization of pectic arabinan and galactan side chains in association with cellulose microfibrils in primary cell walls and related models envisaged. J. Exp. Bot. 2007, 58, 1795–1802. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Popper, Z.A.; Fry, S.C. Widespread occurrence of a covalent linkage between xyloglucan and acidic polysaccharides in suspension-cultured angiosperm cells. Ann. Bot. 2005, 96, 9199. [Google Scholar] [CrossRef] [Green Version]
- Moore, J.; Farrant, J.; Driouich, A. A role for pectin-associated arabinans in maintaining the flexibility of the plant cell wall during water deficit stress. Plant Signal. Behav. 2008, 3, 102–104. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, C.; Zayed, O.; Zeng, F.; Liu, C.; Zhang, L.; Zhu, P.; Hsu, C.C.; Tuncil, Y.E.; Tao, W.A.; Carpita, N.C.; et al. Arabinose biosynthesis is critical for salt stress tolerance in Arabidopsis. New Phytol. 2019, 224, 274–290. [Google Scholar] [CrossRef] [PubMed]
- Nguema-Ona, E.; Vicré-Gibouin, M.; Cannesan, M.A.; Driouich, A. Arabinogalactan proteins in root-microbe interactions. Trends Plant Sci. 2013, 18, 1360–1385. [Google Scholar] [CrossRef]
- Mareri, L.; Romi, M.; Cai, G. Arabinogalactan proteins: Actors or spectators during abiotic and biotic stress in plants? Plant Biosyst. Int. J. Deal. Asp. Plant Biol. 2019, 153, 173–185. [Google Scholar] [CrossRef]
- Nguema-Ona, E.; Moore, J.P.; Fagerström, A.D.; Fangel, J.U.; Willats, W.G.T.; Hugo, A.; Vivier, M.A. Overexpression of the grapevine PGIP1 in tobacco results in compositional changes in the leaf arabinoxyloglucan network in the absence of fungal infection. BMC Plant Biol. 2013, 13, 46. [Google Scholar] [CrossRef]
- Li, Y.L.; Yu, Y.K.; Zhu, K.M.; Ding, L.-N.; Wang, Z.; Yang, Y.-H.; Cao, J.; Xu, L.-Z.; Li, Y.-M.; Tan, X.-L. Down-regulation of MANNANASE7 gene in Brassica napus L. enhances silique dehiscence-resistance. Plant Cell Rep. 2021, 40, 361–374. [Google Scholar] [CrossRef] [PubMed]
- Lahlali, R.; Kumar, S.; Wang, L.; Forseille, L.; Sylvain, N.; Korbas, M.; Muir, D.; Swerhone, G.; Lawrence, J.R.; Fobert, P.R.; et al. Cell wall biomolecular composition plays a potential role in the host type ii resistance to Fusarium head blight in wheat. Front. Microbiol. 2016, 7, 910. [Google Scholar] [CrossRef]
- Kacurakova, M.; Capek, P.; Sasinkova, V.; Wellner, N.; Ebringerova, A. FT-IR study of plant cell wall model compound: Pectic polysaccharides and hemicelluloses. Carbohydr. Polym. 2000, 43, 195–203. [Google Scholar] [CrossRef]
- Cosgrove, D. Microbial expansins. Annu. Rev. Microbiol. 2017, 71, 479–497. [Google Scholar] [CrossRef] [PubMed]







| Code | Specificity | Group |
|---|---|---|
| JIM5 | Binds to partially methyl esterified HG | HG |
| JIM7 | Binds to heavily methyl esterified HG | |
| LM18 | Binds to de-esterified HG, higher affinity to shorter chain (DP < 4) | |
| LM19 | Binds to de-esterified HG, higher affinity to longer chain (DP > 4) | |
| LM20 | Binds to methyl-esterified HG | |
| LM8 | Binds to xylogalacturonan | |
| INRA-RU1 | Binds to unbranched region of RGI, need >6 disaccharide backbone repeats, maximum binding to DP = 14 | RGI and side chains |
| INRA-RU2 | Binds to unbranched region of RGI, significant binding to 2 disaccharide backbone repeats, need at least DP = 4 | |
| LM5 | Binds to (1–4)-ß-D-galactan | |
| LM6 | Binds to (1,5)-α-L-arabinan, may bind to AGP | |
| LM13 | Binds to linear arabinan, highly sensitive to arabinanase | |
| LM16 | Binds to galactan stub | |
| LM21 | binds to mannans, glucomannans, and galactomannans. Binds most strongly to mannotetraose and mannopentaose. | Hemicellulose |
| BS-400-2 | Binds to (1–3)-ß-D-glucan | |
| LM15 | Binds to xyloglucan, XXXG motif | |
| LM24 | Binds to XLLG or XXLG | |
| LM25 | Binds to xyloglucan/unsubstituted glucan, | |
| LM10 | (1–4)-ß-D-xylan | |
| LM11 | bind to unsubstituted xylans and arabinoxylans carrying a low degree of arabinose substitution | |
| CBM3a | Binds to crystalline cellulose and can detect cellulose in both in vitro assays and directly in plant materials | Cellulose |
| JIM8 | Arabinogalactan protein | Glycoprotein |
| JIM13 | Arabinogalactan protein | |
| JIM15 | Arabinogalactan protein | |
| JIM16 | Arabinogalactan protein | |
| JIM17 | Arabinogalactan protein | |
| LM2 | Arabinogalactan protein | |
| MAC207 | Arabinogalactan protein |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gao, Y.; Yin, X.; Jiang, H.; Hansen, J.; Jørgensen, B.; Moore, J.P.; Fu, P.; Wu, W.; Yang, B.; Ye, W.; et al. Comprehensive Leaf Cell Wall Analysis Using Carbohydrate Microarrays Reveals Polysaccharide-Level Variation between Vitis Species with Differing Resistance to Downy Mildew. Polymers 2021, 13, 1379. https://doi.org/10.3390/polym13091379
Gao Y, Yin X, Jiang H, Hansen J, Jørgensen B, Moore JP, Fu P, Wu W, Yang B, Ye W, et al. Comprehensive Leaf Cell Wall Analysis Using Carbohydrate Microarrays Reveals Polysaccharide-Level Variation between Vitis Species with Differing Resistance to Downy Mildew. Polymers. 2021; 13(9):1379. https://doi.org/10.3390/polym13091379
Chicago/Turabian StyleGao, Yu, Xiangjing Yin, Haoyu Jiang, Jeanett Hansen, Bodil Jørgensen, John P. Moore, Peining Fu, Wei Wu, Bohan Yang, Wenxiu Ye, and et al. 2021. "Comprehensive Leaf Cell Wall Analysis Using Carbohydrate Microarrays Reveals Polysaccharide-Level Variation between Vitis Species with Differing Resistance to Downy Mildew" Polymers 13, no. 9: 1379. https://doi.org/10.3390/polym13091379
APA StyleGao, Y., Yin, X., Jiang, H., Hansen, J., Jørgensen, B., Moore, J. P., Fu, P., Wu, W., Yang, B., Ye, W., Song, S., & Lu, J. (2021). Comprehensive Leaf Cell Wall Analysis Using Carbohydrate Microarrays Reveals Polysaccharide-Level Variation between Vitis Species with Differing Resistance to Downy Mildew. Polymers, 13(9), 1379. https://doi.org/10.3390/polym13091379