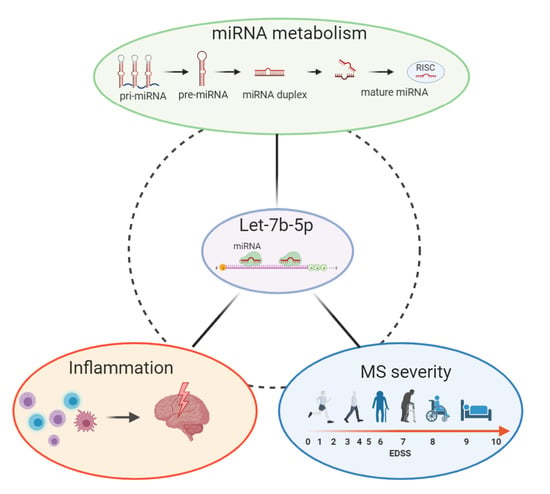
The microRNA let-7b-5p Is Negatively Associated with Inflammation and Disease Severity in Multiple Sclerosis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Let-7 Target mRNA Analysis and Gene Ontology Enrichment Analysis
2.2. Clinical Study Design
2.3. Patients with MS
Clinical Parameters
2.4. RNA Extraction from CSF and miRNA Detection
2.5. Detection of Inflammation-Related Protein in the CSF
2.6. Statistical Analysis
3. Results
3.1. The Let-7 Family Regulates Crucial Processes Involved in MS Pathophysiology
3.2. Let-7b-5p Is a Possible Regulatory Hub of the Pattern of MS-Related miRNAs Circulating in the CSF
3.3. Let-7b-5p Is a Putative Anti-Inflammatory Regulator of the Complex Pathway of Soluble Factors Circulating in the MS CSF
3.4. The miR Let-7b-5p Is Reduced in the CSF of Patients with Progressive MS and Is Associated with Different Processes According to the Phase of the Disease
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Dendrou, C.A.; Fugger, L.; Friese, M.A. Immunopathology of multiple sclerosis. Nat. Rev. Immunol. 2015, 15, 545–558. [Google Scholar] [CrossRef] [PubMed]
- Kunkl, M.; Frascolla, S.; Amormino, C.; Volpe, E.; Tuosto, L. T Helper Cells: The Modulators of Inflammation in Multiple Sclerosis. Cells 2020, 9, 482. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Compston, A.; Coles, A. Multiple sclerosis. Lancet 2008, 372, 1502–1517. [Google Scholar] [CrossRef]
- Harris, V.K.; Tuddenham, J.F.; Sadiq, S.A. Biomarkers of multiple sclerosis: Current findings. Degener. Neurol. Neuromuscul. Dis. 2017, 7, 19–29. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martinez, B.; Peplow, P.V. MicroRNAs in blood and cerebrospinal fluid as diagnostic biomarkers of multiple sclerosis and to monitor disease progression. Neural Regen. Res. 2020, 15, 606–619. [Google Scholar] [CrossRef]
- Perdaens, O.; Dang, H.A.; D’Auria, L.; van Pesch, V. CSF microRNAs discriminate MS activity and share similarity to other neuroinflammatory disorders. Neurol. Neuroimmunol. Neuroinflamm. 2020, 7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krol, J.; Loedige, I.; Filipowicz, W. The widespread regulation of microRNA biogenesis, function and decay. Nat. Rev. Genet. 2010, 11, 597–610. [Google Scholar] [CrossRef]
- Friedman, R.C.; Farh, K.K.-H.; Burge, C.B.; Bartel, D.P. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009, 19, 92–105. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vidigal, J.A.; Ventura, A. The biological functions of miRNAs: Lessons from in vivo studies. Trends Cell Biol. 2015, 25, 137–147. [Google Scholar] [CrossRef] [Green Version]
- Ivey, K.N.; Srivastava, D. MicroRNAs as Developmental Regulators. Cold Spring Harb. Perspect. Biol. 2015, 7, a008144. [Google Scholar] [CrossRef] [Green Version]
- Long, H.; Wang, X.; Chen, Y.; Wang, L.; Zhao, M.; Lu, Q. Dysregulation of microRNAs in autoimmune diseases: Pathogenesis, biomarkers and potential therapeutic targets. Cancer Lett. 2018, 428, 90–103. [Google Scholar] [CrossRef] [PubMed]
- Angelou, C.C.; Wells, A.C.; Vijayaraghavan, J.; Dougan, C.E.; Lawlor, R.; Iverson, E.; Lazarevic, V.; Kimura, M.Y.; Pobezinsky, L.A. Differentiation of Pathogenic Th17 Cells Is Negatively Regulated by Let-7 MicroRNAs in a Mouse Model of Multiple Sclerosis. Front. Immunol. 2020, 10, 3125. [Google Scholar] [CrossRef] [Green Version]
- Kimura, K.; Hohjoh, H.; Fukuoka, M.; Sato, W.; Oki, S.; Tomi, C.; Yamaguchi, H.; Kondo, T.; Takahashi, R.; Yamamura, T. Circulating exosomes suppress the induction of regulatory T cells via let-7i in multiple sclerosis. Nat. Commun. 2018, 9, 17. [Google Scholar] [CrossRef]
- Roush, S.; Slack, F.J. The let-7 family of microRNAs. Trends Cell Biol. 2008, 18, 505–516. [Google Scholar] [CrossRef]
- Guan, H.; Fan, D.; Mrelashvili, D.; Hao, H.; Singh, N.P.; Singh, U.P.; Nagarkatti, P.S.; Nagarkatti, M. MicroRNA let-7e is associated with the pathogenesis of experimental autoimmune encephalomyelitis. Eur. J. Immunol. 2013, 43, 104–114. [Google Scholar] [CrossRef] [PubMed]
- Lehmann, S.M.; Krüger, C.; Park, B.; Derkow, K.; Rosenberger, K.; Baumgart, J.; Trimbuch, T.; Eom, G.; Hinz, M.; Kaul, D.; et al. An unconventional role for miRNA: Let-7 activates Toll-like receptor 7 and causes neurodegeneration. Nat. Neurosci. 2012, 15, 827–835. [Google Scholar] [CrossRef] [PubMed]
- Gaudet, A.D.; Fonken, L.K.; Watkins, L.R.; Nelson, R.J.; Popovich, P.G. MicroRNAs: Roles in Regulating Neuroinflammation. Neurosci. Rev. J. Bringing Neurobiol. Neurol. Psychiatry 2018, 24, 221–245. [Google Scholar] [CrossRef] [Green Version]
- Liguori, M.; Nuzziello, N.; Licciulli, F.; Consiglio, A.; Simone, M.; Viterbo, R.G.; Creanza, T.M.; Ancona, N.; Tortorella, C.; Margari, L.; et al. Combined microRNA and mRNA expression analysis in pediatric multiple sclerosis: An integrated approach to uncover novel pathogenic mechanisms of the disease. Hum. Mol. Genet. 2018, 27, 66–79. [Google Scholar] [CrossRef]
- Manna, I.; Iaccino, E.; Dattilo, V.; Barone, S.; Vecchio, E.; Mimmi, S.; Filippelli, E.; Demonte, G.; Polidoro, S.; Granata, A.; et al. Exosome-associated miRNA profile as a prognostic tool for therapy response monitoring in multiple sclerosis patients. FASEB J. 2018, 32, 4241–4246. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Licursi, V.; Conte, F.; Fiscon, G.; Paci, P. MIENTURNET: An interactive web tool for microRNA-target enrichment and network-based analysis. BMC Bioinform. 2019, 20, 545. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, H.-Y.; Lin, Y.-C.-D.; Li, J.; Huang, K.-Y.; Shrestha, S.; Hong, H.-C.; Tang, Y.; Chen, Y.-G.; Jin, C.-N.; Yu, Y.; et al. miRTarBase 2020: Updates to the experimentally validated microRNA-target interaction database. Nucleic Acids Res. 2020, 48, D148–D154. [Google Scholar] [CrossRef] [Green Version]
- Yu, G.; Wang, L.-G.; Han, Y.; He, Q.-Y. ClusterProfiler: An R package for comparing biological themes among gene clusters. OMICS 2012, 16, 284–287. [Google Scholar] [CrossRef] [PubMed]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef] [Green Version]
- Mandolesi, G.; Gentile, A.; Musella, A.; Fresegna, D.; De Vito, F.; Bullitta, S.; Sepman, H.; Marfia, G.A.; Centonze, D. Synaptopathy connects inflammation and neurodegeneration in multiple sclerosis. Nat. Rev. Neurol. 2015, 11, 711–724. [Google Scholar] [CrossRef] [PubMed]
- McGowan, H.; Mirabella, V.R.; Hamod, A.; Karakhanyan, A.; Mlynaryk, N.; Moore, J.C.; Tischfield, J.A.; Hart, R.P.; Pang, Z.P. hsa-let-7c miRNA Regulates Synaptic and Neuronal Function in Human Neurons. Front. Synaptic Neurosci. 2018, 10, 19. [Google Scholar] [CrossRef]
- Gandhi, R. miRNA in multiple sclerosis: Search for novel biomarkers. Mult. Scler. 2015, 21, 1095–1103. [Google Scholar] [CrossRef] [Green Version]
- Huang, Q.; Xiao, B.; Ma, X.; Qu, M.; Li, Y.; Nagarkatti, P.; Nagarkatti, M.; Zhou, J. MicroRNAs associated with the pathogenesis of multiple sclerosis. J. Neuroimmunol. 2016, 295–296, 148–161. [Google Scholar] [CrossRef]
- Mandolesi, G.; De Vito, F.; Musella, A.; Gentile, A.; Bullitta, S.; Fresegna, D.; Sepman, H.; Di Sanza, C.; Haji, N.; Mori, F.; et al. miR-142-3p Is a Key Regulator of IL-1β-Dependent Synaptopathy in Neuroinflammation. J. Neurosci. 2017, 37, 546–561. [Google Scholar] [CrossRef]
- Polman, C.H.; Reingold, S.C.; Banwell, B.; Clanet, M.; Cohen, J.A.; Filippi, M.; Fujihara, K.; Havrdova, E.; Hutchinson, M.; Kappos, L.; et al. Diagnostic criteria for multiple sclerosis: 2010 revisions to the McDonald criteria. Ann. Neurol. 2011, 69, 292–302. [Google Scholar] [CrossRef] [Green Version]
- Stampanoni Bassi, M.; Buttari, F.; Simonelli, I.; Gilio, L.; Furlan, R.; Finardi, A.; Marfia, G.A.; Visconti, A.; Paolillo, A.; Storto, M.; et al. A Single Nucleotide ADA Genetic Variant Is Associated to Central Inflammation and Clinical Presentation in MS: Implications for Cladribine Treatment. Genes 2020, 11, 1152. [Google Scholar] [CrossRef]
- Costa, A.; Bagoj, E.; Monaco, M.; Zabberoni, S.; De Rosa, S.; Papantonio, A.M.; Mundi, C.; Caltagirone, C.; Carlesimo, G.A. Standardization and normative data obtained in the Italian population for a new verbal fluency instrument, the phonemic/semantic alternate fluency test. Neurol. Sci. 2014, 35, 365–372. [Google Scholar] [CrossRef] [PubMed]
- Measso, G.; Cavarzeran, F.; Zappalà, G.; Lebowitz, B.D.; Crook, T.H.; Pirozzolo, F.J.; Amaducci, L.A.; Massari, D.; Grigoletto, F. The Mini-Mental State Examination: Normative Study of An Italian Random Sample. Dev. Neuropsychol. 1993, 9, 77–85. [Google Scholar] [CrossRef]
- Vandesompele, J.; De Preter, K.; Pattyn, F.; Poppe, B.; Van Roy, N.; De Paepe, A.; Speleman, F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002, 3, research0034.1. [Google Scholar] [CrossRef] [Green Version]
- Marabita, F.; de Candia, P.; Torri, A.; Tegnér, J.; Abrignani, S.; Rossi, R.L. Normalization of circulating microRNA expression data obtained by quantitative real-time RT-PCR. Brief. Bioinform. 2016, 17, 204–212. [Google Scholar] [CrossRef] [Green Version]
- Bergman, P.; Piket, E.; Khademi, M.; James, T.; Brundin, L.; Olsson, T.; Piehl, F.; Jagodic, M. Circulating miR-150 in CSF is a novel candidate biomarker for multiple sclerosis. Neurol. Neuroimmunol. Neuroinflamm. 2016, 3, e219. [Google Scholar] [CrossRef] [Green Version]
- Bruinsma, I.B.; van Dijk, M.; Bridel, C.; van de Lisdonk, T.; Haverkort, S.Q.; Runia, T.F.; Steinman, L.; Hintzen, R.Q.; Killestein, J.; Verbeek, M.M.; et al. Regulator of oligodendrocyte maturation, miR-219, a potential biomarker for MS. J. Neuroinflamm. 2017, 14, 235. [Google Scholar] [CrossRef]
- Gallego, J.A.; Gordon, M.L.; Claycomb, K.; Bhatt, M.; Lencz, T.; Malhotra, A.K. In vivo microRNA detection and quantitation in cerebrospinal fluid. J. Mol. Neurosci. 2012, 47, 243–248. [Google Scholar] [CrossRef] [Green Version]
- Burgos, K.L.; Javaherian, A.; Bomprezzi, R.; Ghaffari, L.; Rhodes, S.; Courtright, A.; Tembe, W.; Kim, S.; Metpally, R.; Van Keuren-Jensen, K. Identification of extracellular miRNA in human cerebrospinal fluid by next-generation sequencing. RNA 2013, 19, 712–722. [Google Scholar] [CrossRef] [Green Version]
- Harris, V.K.; Sadiq, S.A. Biomarkers of therapeutic response in multiple sclerosis: Current status. Mol. Diagn. Ther. 2014, 18, 605–617. [Google Scholar] [CrossRef] [Green Version]
- Stoicea, N.; Du, A.; Lakis, D.C.; Tipton, C.; Arias-Morales, C.E.; Bergese, S.D. The MiRNA Journey from Theory to Practice as a CNS Biomarker. Front. Genet. 2016, 7, 11. [Google Scholar] [CrossRef] [Green Version]
- Lescher, J.; Paap, F.; Schultz, V.; Redenbach, L.; Scheidt, U.; Rosewich, H.; Nessler, S.; Fuchs, E.; Gärtner, J.; Brück, W.; et al. MicroRNA regulation in experimental autoimmune encephalomyelitis in mice and marmosets resembles regulation in human multiple sclerosis lesions. J. Neuroimmunol. 2012, 246, 27–33. [Google Scholar] [CrossRef] [PubMed]
- Thamilarasan, M.; Koczan, D.; Hecker, M.; Paap, B.; Zettl, U.K. MicroRNAs in multiple sclerosis and experimental autoimmune encephalomyelitis. Autoimmun. Rev. 2012, 11, 174–179. [Google Scholar] [CrossRef]
- Freiesleben, S.; Hecker, M.; Zettl, U.K.; Fuellen, G.; Taher, L. Analysis of microRNA and Gene Expression Profiles in Multiple Sclerosis: Integrating Interaction Data to Uncover Regulatory Mechanisms. Sci. Rep. 2016, 6, 34512. [Google Scholar] [CrossRef]
- Wei, T.; Simko, V.; Levy, M.; Xie, Y.; Jin, Y.; Zemla, J. Package “Corrplot”. 2017. Available online: https://cran.r-project.org/web/packages/corrplot/corrplot.pdf (accessed on 2 February 2021).
- Hastie, T.; Tibshirani, R.; Friedman, J. The Elements of Statistical Learning: Data Mining, Inference, and Prediction; Springer Science & Business Media: Berlin, Germany, 2009. [Google Scholar]
- Csardi, G.; Nepusz, T. The igraph software package for complex network research. Int. J. Commun. Syst. 2006, 1695, 1–9. [Google Scholar]
- Kaufman, L.; Rousseeuw, P.J. Finding Groups in Data: An Introduction to Cluster Analysis; John Wiley & Sons: Hoboken, NJ, USA, 2009. [Google Scholar]
- Langfelder, P.; Horvath, S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinform. 2008, 9, 559. [Google Scholar] [CrossRef] [Green Version]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Griffiths-Jones, S. The microRNA Registry. Nucleic Acids Res. 2004, 32, D109–D111. [Google Scholar] [CrossRef]
- Olsson, T.; Barcellos, L.F.; Alfredsson, L. Interactions between genetic, lifestyle and environmental risk factors for multiple sclerosis. Nat. Rev. Neurol. 2017, 13, 25–36. [Google Scholar] [CrossRef] [PubMed]
- Munõz-San Martín, M.; Reverter, G.; Robles-Cedenõ, R.; Buxò, M.; Ortega, F.J.; Gómez, I.; Tomàs-Roig, J.; Celarain, N.; Villar, L.M.; Perkal, H.; et al. Analysis of miRNA signatures in CSF identifies upregulation of miR-21 and miR-146a/b in patients with multiple sclerosis and active lesions. J. Neuroinflamm. 2019, 16, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Haghikia, A.; Hellwig, K.; Baraniskin, A.; Holzmann, A.; Décard, B.F.; Thum, T. Regulated microRNAs in the CSF of patients with multiple sclerosis. Neurology 2012, 79, 2166–2170. [Google Scholar] [CrossRef]
- Ebrahimkhani, S.; Vafaee, F.; Young, P.E.; Hur, S.S.J.; Hawke, S.; Devenney, E.; Beadnall, H.; Barnett, M.H.; Suter, C.M.; Buckland, M.E. Exosomal microRNA signatures in multiple sclerosis reflect disease status. Sci. Rep. 2017, 7, 14293. [Google Scholar] [CrossRef] [Green Version]
- Juźwik, C.A.; Drake, S.; Zhang, Y.; Paradis-Isler, N.; Sylvester, A.; Amar-Zifkin, A.; Douglas, C.; Morquette, B.; Moore, C.S.; Fournier, A.E. MicroRNA dysregulation in neurodegenerative diseases: A systematic review. Prog. Neurobiol. 2019, 182, 101664. [Google Scholar] [CrossRef]
- Cantoni, C.; Cignarella, F.; Ghezzi, L.; Mikesell, B.; Bollman, B.; Berrien-Elliott, M.M.; Ireland, A.R.; Fehniger, T.A.; Wu, G.F.; Piccio, L. Mir-223 regulates the number and function of myeloid-derived suppressor cells in multiple sclerosis and experimental autoimmune encephalomyelitis. Acta Neuropathol. 2017, 133, 61–77. [Google Scholar] [CrossRef]
- Finardi, A.; Diceglie, M.; Carbone, L.; Arnò, C.; Mandelli, A.; De Santis, G.; Fedeli, M.; Dellabona, P.; Casorati, G.; Furlan, R. Mir106b-25 and Mir17-92 Are Crucially Involved in the Development of Experimental Neuroinflammation. Front. Neurol. 2020, 11, 912. [Google Scholar] [CrossRef]
- Keller, A.; Leidinger, P.; Steinmeyer, F.; Stähler, C.; Franke, A.; Hemmrich-Stanisak, G.; Kappel, A.; Wright, I.; Dörr, J.; Paul, F.; et al. Comprehensive analysis of microRNA profiles in multiple sclerosis including next-generation sequencing. Mult. Scler. 2014, 20, 295–303. [Google Scholar] [CrossRef]
- Arruda, L.C.M.; Lorenzi, J.C.C.; Sousa, A.P.A.; Zanette, D.L.; Palma, P.V.B.; Panepucci, R.A.; Brum, D.S.; Barreira, A.A.; Covas, D.T.; Simões, B.P.; et al. Autologous hematopoietic SCT normalizes miR-16, -155 and -142-3p expression in multiple sclerosis patients. Bone Marrow Transplant. 2015, 50, 380–389. [Google Scholar] [CrossRef]
- Sun, X.; Zhang, H. miR-451 elevation relieves inflammatory pain by suppressing microglial activation-evoked inflammatory response via targeting TLR4. Cell Tissue Res. 2018, 374, 487–495. [Google Scholar] [CrossRef]
- Morquette, B.; Juźwik, C.A.; Drake, S.S.; Charabati, M.; Zhang, Y.; Lécuyer, M.-A.; Galloway, D.A.; Dumas, A.; de Faria Junior, O.; Paradis-Isler, N.; et al. MicroRNA-223 protects neurons from degeneration in experimental autoimmune encephalomyelitis. Brain 2019, 142, 2979–2995. [Google Scholar] [CrossRef]
- Teuber-Hanselmann, S.; Meinl, E.; Junker, A. MicroRNAs in gray and white matter multiple sclerosis lesions: Impact on pathophysiology. J. Pathol. 2020, 250, 496–509. [Google Scholar] [CrossRef] [Green Version]
- Letellier, M.; Elramah, S.; Mondin, M.; Soula, A.; Penn, A.; Choquet, D.; Landry, M.; Thoumine, O.; Favereaux, A. miR-92a regulates expression of synaptic GluA1-containing AMPA receptors during homeostatic scaling. Nat. Neurosci. 2014, 17, 1040–1042. [Google Scholar] [CrossRef] [PubMed]
- Junker, A.; Krumbholz, M.; Eisele, S.; Mohan, H.; Augstein, F.; Bittner, R.; Lassmann, H.; Wekerle, H.; Hohlfeld, R.; Meinl, E. MicroRNA profiling of multiple sclerosis lesions identifies modulators of the regulatory protein CD47. Brain 2009, 132, 3342–3352. [Google Scholar] [CrossRef] [Green Version]
- Iliopoulos, D.; Hirsch, H.A.; Struhl, K. An epigenetic switch involving NF-kappaB, Lin28, Let-7 MicroRNA, and IL6 links inflammation to cell transformation. Cell 2009, 139, 693–706. [Google Scholar] [CrossRef] [Green Version]
- Sung, S.-Y.; Liao, C.-H.; Wu, H.-P.; Hsiao, W.-C.; Wu, I.-H.; Yu, J.; Lin, S.-H.; Hsieh, C.-L. Loss of let-7 microRNA upregulates IL-6 in bone marrow-derived mesenchymal stem cells triggering a reactive stromal response to prostate cancer. PLoS ONE 2013, 8, e71637. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gong, Z.; Zhao, S.; Zhang, J.; Xu, X.; Guan, W.; Jing, L.; Liu, P.; Lu, J.; Teng, J.; Peng, T.; et al. Initial research on the relationship between let-7 family members in the serum and massive cerebral infarction. J. Neurol. Sci. 2016, 361, 150–157. [Google Scholar] [CrossRef]
- Wang, X.; Wang, H.-X.; Li, Y.-L.; Zhang, C.-C.; Zhou, C.-Y.; Wang, L.; Xia, Y.-L.; Du, J.; Li, H.-H. MicroRNA Let-7i negatively regulates cardiac inflammation and fibrosis. Hypertension 2015, 66, 776–785. [Google Scholar] [CrossRef]
- Jiang, L.; Cheng, Z.; Qiu, S.; Que, Z.; Bao, W.; Jiang, C.; Zou, F.; Liu, P.; Liu, J. Altered let-7 expression in Myasthenia gravis and let-7c mediated regulation of IL-10 by directly targeting IL-10 in Jurkat cells. Int. Immunopharmacol. 2012, 14, 217–223. [Google Scholar] [CrossRef]
- Filiano, A.J.; Gadani, S.P.; Kipnis, J. How and why do T cells and their derived cytokines affect the injured and healthy brain? Nat. Rev. Neurosci. 2017, 18, 375–384. [Google Scholar] [CrossRef] [Green Version]
- Gentile, A.; De Vito, F.; Fresegna, D.; Rizzo, F.R.; Bullitta, S.; Guadalupi, L.; Vanni, V.; Buttari, F.; Stampanoni Bassi, M.; Leuti, A.; et al. Peripheral T cells from multiple sclerosis patients trigger synaptotoxic alterations in central neurons. Neuropathol. Appl. Neurobiol. 2020, 46, 160–170. [Google Scholar] [CrossRef]
- Tufekci, K.U.; Oner, M.G.; Genc, S.; Genc, K. MicroRNAs and Multiple Sclerosis. Autoimmune Dis. 2010, 2011, 807426. [Google Scholar] [CrossRef] [Green Version]
- Nuzziello, N.; Ciaccia, L.; Liguori, M. Precision Medicine in Neurodegenerative Diseases: Some Promising Tips Coming from the microRNAs’ World. Cells 2020, 9, 75. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, Y.; Peng, R.; Peng, H.; Liu, H.; Wen, L.; Wu, T.; Yi, H.; Li, A.; Zhang, Z. miR-451 suppresses the NF-kappaB-mediated proinflammatory molecules expression through inhibiting LMP7 in diabetic nephropathy. Mol. Cell. Endocrinol. 2016, 433, 75–86. [Google Scholar] [CrossRef]
- Teng, G.; Wang, W.; Dai, Y.; Wang, S.; Chu, Y.; Li, J. Let-7b is involved in the inflammation and immune responses associated with Helicobacter pylori infection by targeting Toll-like receptor 4. PLoS ONE 2013, 8, e56709. [Google Scholar] [CrossRef] [Green Version]
- Essandoh, K.; Li, Y.; Huo, J.; Fan, G.-C. MiRNA-Mediated Macrophage Polarization and its Potential Role in the Regulation of Inflammatory Response. Shock 2016, 46, 122–131. [Google Scholar] [CrossRef]
- Ghadiri, N.; Emamnia, N.; Ganjalikhani-Hakemi, M.; Ghaedi, K.; Etemadifar, M.; Salehi, M.; Shirzad, H.; Nasr-Esfahani, M.H. Analysis of the expression of mir-34a, mir-199a, mir-30c and mir-19a in peripheral blood CD4+T lymphocytes of relapsing-remitting multiple sclerosis patients. Gene 2018, 659, 109–117. [Google Scholar] [CrossRef]
- Vistbakka, J.; Sumelahti, M.-L.; Lehtimäki, T.; Elovaara, I.; Hagman, S. Evaluation of serum miR-191-5p, miR-24-3p, miR-128-3p, and miR-376c-3 in multiple sclerosis patients. Acta Neurol. Scand. 2018, 138, 130–136. [Google Scholar] [CrossRef]
- Marques-Rocha, J.L.; Garcia-Lacarte, M.; Samblas, M.; Bressan, J.; Martínez, J.A.; Milagro, F.I. Regulatory roles of miR-155 and let-7b on the expression of inflammation-related genes in THP-1 cells: Effects of fatty acids. J. Physiol. Biochem. 2018, 74, 579–589. [Google Scholar] [CrossRef]
- Zhao, C.; Sun, G.; Li, S.; Lang, M.-F.; Yang, S.; Li, W.; Shi, Y. MicroRNA let-7b regulates neural stem cell proliferation and differentiation by targeting nuclear receptor TLX signaling. Proc. Natl. Acad. Sci. USA 2010, 107, 1876–1881. [Google Scholar] [CrossRef] [Green Version]
- Schulte, L.N.; Eulalio, A.; Mollenkopf, H.-J.; Reinhardt, R.; Vogel, J. Analysis of the host microRNA response to Salmonella uncovers the control of major cytokines by the let-7 family. EMBO J. 2011, 30, 1977–1989. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Salvi, V.; Gianello, V.; Tiberio, L.; Sozzani, S.; Bosisio, D. Cytokine Targeting by miRNAs in Autoimmune Diseases. Front. Immunol. 2019, 10, 15. [Google Scholar] [CrossRef]







| Patients’ Group | |||||
|---|---|---|---|---|---|
| Variable | Control Subjects | All Patients | CIS/RIS | RRMS | PMS |
| N | 20 | 166 | 25 | 117 | 24 |
| Age | 46.1 (29.7–53.3) | 39.8 (29.7–47.9) | 41.0 (32.9–42.9) | 35.6 (27.7–46.7) | 48.6 (39.6–55.5) |
| Gender: F | 7 (35.0%) | 57 (34.3%) | 6 (24%) | 38 (32.5%) | 13 (54.2%) |
| Oligoclonal Bandingy/n/NA | / | 27 (17.2%) | 10/14/1 (41.7%) | 15/95/7 (13.6%) | 2/21/1 (8.7%) |
| Disease activityy/n/NA | / | 81 (48.8%) | 10/14/1 (41.7%) | 67/43/7 (60.9%) | 4 (16.7%) |
| EDSS | / | 2.0 (1.0–3.0) | 1.0 (1.0–2.0) | 1.5 (1.0–3.0) | 3.5 (2.2–5.2) |
| Disease Duration | / | 12.4 (2.4–37.7) | 2.8 (1.6–4.7) | 12.9 (1.8–38.9) | 24.3 (12.3–61.3) |
| PI (T0) | / | 0.2 (0.0–0.6) | 0.6 (0.2–1.0) | 0.1 (0.0–0.6) | 0.2 (0.0–0.2) |
| let-7b-5p | 0.002 (0.001–0.009) | 0.004 (0.001–0.012) | 0.005 (0.001–0.010) | 0.005 (0.001–0.015) | 0.002 (0.001–0.003) |
| let-7e-5p | 0.003 (0.001–0.010) | 0.002 (0.001–0.006) | 0.002 (0.000–0.01) | 0.002 (0.001–0.006) | 0.004 (0.002–0.006) |
| let-7f-5p | 0.004 (0.003–0.007) | 0.004 (0.002–0.006) | 0.004 (0.003–0.007) | 0.004 (0.002–0.006) | 0.004 (0.002–0.005) |
| corr | p Value | p Adjusted | |||
|---|---|---|---|---|---|
| Cytokines | IFNγ | 0.224 | 0.004 | 0.006 | Cluster 1 |
| IL1ra | 0.311 | <0.001 | <0.001 | ||
| IL5 | 0.308 | <0.001 | 0.0002 | ||
| IL2 | −0.386 | <0.001 | <0.001 | Cluster 2 | |
| IL6 | −0.277 | <0.001 | 0.001 | ||
| IL10 | −0.257 | 0.001 | 0.002 | ||
| IL12_p70 | −0.294 | <0.001 | <0.001 | ||
| IL15 | −0.348 | <0.001 | <0.001 | ||
| IL17 | −0.247 | 0.001 | 0.002 | ||
| GM_CSF | −0.420 | <0.001 | <0.001 | ||
| Chemokines | IL8 | 0.311 | <0.001 | <0.001 | Cluster 1 |
| IP10 | 0.379 | <0.001 | <0.001 | ||
| Rantes | 0.392 | <0.001 | <0.001 | ||
| Eotaxin | −0.234 | 0.002 | 0.004 | Cluster 2 | |
| MIP1b | −0.237 | 0.002 | 0.004 | ||
| Growth Factors | G_CSF | 0.387 | <0.001 | <0.001 | Cluster 1 |
| bFGF | −0.340 | <0.001 | <0.001 | Cluster 2 | |
| PDGF bb | −0.307 | <0.001 | <0.001 |
| corr | p Value | p Adjusted | |||
|---|---|---|---|---|---|
| Cytokines | IFNγ | 0.249 | 0.003 | 0.005 | Cluster 1 |
| IL1ra | 0.317 | <0.001 | <0.001 | ||
| IL5 | 0.287 | 0.001 | 0.001 | ||
| IL2 | −0.383 | <0.001 | <0.001 | Cluster 2 | |
| IL6 | −0.293 | <0.001 | 0.001 | ||
| IL10 | −0.266 | 0.001 | 0.003 | ||
| IL12_p70 | −0.283 | 0.001 | 0.001 | ||
| IL15 | −0.338 | <0.001 | <0.001 | ||
| IL17 | −0.248 | 0.003 | 0.005 | ||
| GM_CSF | −0.416 | <0.001 | <0.001 | ||
| Chemokines | IL8 | 0.309 | <0.001 | 0.001 | Cluster 1 |
| IP10 | 0.388 | <0.001 | <0.001 | ||
| Rantes | 0.364 | <0.001 | <0.001 | ||
| Eotaxin | −0.231 | 0.006 | 0.009 | Cluster 2 | |
| MIP1b | −0.226 | 0.007 | 0.010 | ||
| Growth Factors | G_CSF | 0.381 | <0.001 | <0.001 | Cluster 1 |
| bFGF | −0.324 | <0.001 | <0.001 | Cluster 2 | |
| PDGF bb | −0.299 | <0.001 | 0.001 |
| corr | p Value | p Adjusted | |||
|---|---|---|---|---|---|
| Cytokines | IL5 | 0.567 | 0.004 | 0.035 | Cluster 1 |
| Chemokines | Rantes | 0.628 | 0.001 | 0.028 | |
| Growth Factors | G_CSF | 0.587 | 0.003 | 0.035 |
| Estimate | Std. Error | T-Value | Pr(>|t|) | ||
|---|---|---|---|---|---|
| non-PMS | PC1 cluster 1 | 0.291 −0.233 −0.111 −0.012 −0.185 | 0.010 0.070 0.120 0.012 0.316 | 2.985 −3.306 −0.930 −0.976 −0.587 | 0.003 0.001 0.354 0.331 0.558 |
| PC1 cluster 2 | |||||
| EDSS | |||||
| Age | |||||
| Gender | |||||
| PMS | PC1cluster 1 | 0.177 | 0.087 | 2.045 | 0.057 |
| PC1cluster 2 | −0.042 | 0.099 | −0.427 | 0.674 | |
| EDSS | −0.386 | 0.109 | −3.542 | 0.002 | |
| Age | 0.048 | 0.019 | 2.526 | 0.021 | |
| Gender | −0.143 | 0.347 | −0.412 | 0.685 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mandolesi, G.; Rizzo, F.R.; Balletta, S.; Stampanoni Bassi, M.; Gilio, L.; Guadalupi, L.; Nencini, M.; Moscatelli, A.; Ryan, C.P.; Licursi, V.; et al. The microRNA let-7b-5p Is Negatively Associated with Inflammation and Disease Severity in Multiple Sclerosis. Cells 2021, 10, 330. https://doi.org/10.3390/cells10020330
Mandolesi G, Rizzo FR, Balletta S, Stampanoni Bassi M, Gilio L, Guadalupi L, Nencini M, Moscatelli A, Ryan CP, Licursi V, et al. The microRNA let-7b-5p Is Negatively Associated with Inflammation and Disease Severity in Multiple Sclerosis. Cells. 2021; 10(2):330. https://doi.org/10.3390/cells10020330
Chicago/Turabian StyleMandolesi, Georgia, Francesca Romana Rizzo, Sara Balletta, Mario Stampanoni Bassi, Luana Gilio, Livia Guadalupi, Monica Nencini, Alessandro Moscatelli, Colleen Patricia Ryan, Valerio Licursi, and et al. 2021. "The microRNA let-7b-5p Is Negatively Associated with Inflammation and Disease Severity in Multiple Sclerosis" Cells 10, no. 2: 330. https://doi.org/10.3390/cells10020330
APA StyleMandolesi, G., Rizzo, F. R., Balletta, S., Stampanoni Bassi, M., Gilio, L., Guadalupi, L., Nencini, M., Moscatelli, A., Ryan, C. P., Licursi, V., Dolcetti, E., Musella, A., Gentile, A., Fresegna, D., Bullitta, S., Caioli, S., Vanni, V., Sanna, K., Bruno, A., ... De Vito, F. (2021). The microRNA let-7b-5p Is Negatively Associated with Inflammation and Disease Severity in Multiple Sclerosis. Cells, 10(2), 330. https://doi.org/10.3390/cells10020330