Autophagy-Associated IL-15 Production Is Involved in the Pathogenesis of Leprosy Type 1 Reaction
Abstract
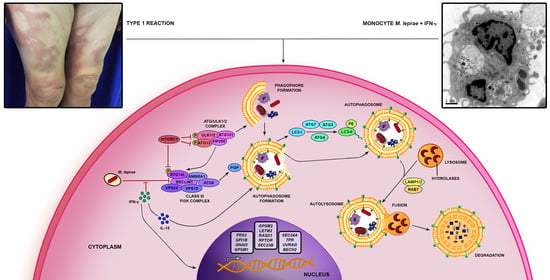
:1. Introduction
2. Materials and Methods
2.1. Patients and Clinical Specimens
2.2. Ethics Statement
2.3. Cell Culture
2.4. Mycobacteria
2.5. Autophagy
2.6. Transmission Electron Microscopy (TEM)
2.7. Immunofluorescence Staining
2.8. Cytokine Measurement
2.9. RNA Isolation, Reverse Transcription, and Real-Time PCR Array Quantifications
2.10. Gene Expression Analysis of IL-15 and IL-10 by Real-Time RT-qPCR
2.11. Autophagy Pathway Analysis
2.12. Leprosy Skin Biopsy Gene Expression from Public Microarray Data
2.13. Western Blot
2.14. Statistical Analysis
3. Results
3.1. IFN-γ Induced the Formation of Autophagosomes in Human Macrophages Stimulated with M. leprae
3.2. IFN-γ Treatment Induced Autophagy Flux in THP-1 Macrophages Stimulated with M. leprae
3.3. IFN-γ Upregulated Autophagy Gene Expression in M. leprae-Stimulated THP-1 Cells
3.4. IFN-γ Increased IL-15 Secretion in THP-1 Macrophages Stimulated with M. leprae
3.5. Proinflammatory Cytokine IL-15 Was Increased in MB Skin Lesions That Progressed to T1R
3.6. Modulation of Autophagy Pathway in T1R Lesions Was Mediated by IFN-γ
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Scollard, D.M.; Martelli, C.M.; Stefani, M.M.; de Fatima Maroja, M.; Villahermosa, L.; Pardillo, F.; Tamang, K.B. Risk factors for leprosy reactions in three endemic countries. Am. J. Trop. Med. Hyg. 2015, 92, 108–114. [Google Scholar] [CrossRef] [Green Version]
- Lockwood, D.N.J.; Lucas, S.B.; Desikan, K.V.; Ebenezer, G.; Suneetha, S.; Nicholls, P. The histological diagnosis of leprosy type 1 reactions: Identification of key variables and an analysis of the process of histological diagnosis. J. Clin. Pathol. 2008, 61, 595–600. [Google Scholar] [CrossRef]
- Naafs, B.; van Hees, C.L.M. Leprosy Type 1 Reaction (Formerly Reversal Reaction). Clin. Dermatol. 2016, 34, 37–50. [Google Scholar] [CrossRef] [PubMed]
- Jih, M.H.; Kimyai-Asadi, A.; Levis, W.R. Reversal reaction to Hansen’s disease. J. Drugs Dermatol. 2002, 1, 70–71. [Google Scholar] [PubMed]
- Smith, W.C.; Anderson, A.M.; Withington, S.G.; van Brakel, W.H.; Croft, R.P.; Nicholls, P.G.; Richardus, J.H. Steroid prophylaxis for prevention of nerve function impairment in leprosy: Randomised placebo controlled trial (TRIPOD 1). BMJ 2004, 328, 1459. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kamath, S.; Vaccaro, S.A.; Rea, T.H.; Ochoa, M.T. Recognizing and managing the immunologic reactions in leprosy. J. Am. Acad. Dermatol. 2014, 71, 795–803. [Google Scholar] [CrossRef]
- Croft, R.P.; Nicholls, P.G.; Steyerberg, E.W.; Richardus, J.H.; Smith, W.C.S. A clinical prediction rule for nerve function impairment in leprosy patients. Lancet 2000, 355, 1603–1606. [Google Scholar] [CrossRef]
- Lockwood, D.N.J.; Nicholls, P.; Smith, W.C.S.; Das, L.; Barkataki, P.; van Brakel, W.; Suneetha, S. Comparing the clinical and histological diagnosis of leprosy and leprosy reactions in the infir cohort of indian patients with multibacillary leprosy. PLoS Negl. Trop. Dis. 2012, 6, e1702. [Google Scholar] [CrossRef] [Green Version]
- Naafs, B. Bangkok Workshop on Leprosy Research. Treatment of reactions and nerve damage. Int. J. Lepr. Other Mycobact. Dis. 1996, 64, S21–S28. [Google Scholar]
- Legendre, D.P.; Muzny, C.A.; Swiatlo, E. Hansen’s Disease (Leprosy): Current and Future Pharmacotherapy and Treatment of Disease-Related Immunologic Reactions. Pharmacotherapy 2012, 32, 27–37. [Google Scholar] [CrossRef]
- Van Veen, N.H.; Nicholls, P.G.; Smith, W.C.; Richardus, J.H. Corticosteroids for treating nerve damage in leprosy. Cochrane Database Syst. Rev. 2016, 23, CD005491. [Google Scholar] [CrossRef]
- Maymone, M.B.C.; Venkatesh, S.; Laughter, M.; Abdat, R.; Hugh, J.; Dacso, M.M.; Dellavalle, R.P. Leprosy: Treatment and Management of Complications. J. Am. Acad. Dermatol. 2020, 83, 17–30. [Google Scholar] [CrossRef]
- Silva, B.J.A.; Barbosa, M.G.M.; Andrade, P.R.; Ferreira, H.; Nery, J.A.C.; Côrte-Real, S.; da Silva, G.M.; Rosa, P.S.; Fabri, M.; Sarno, E.N.; et al. Autophagy Is an Innate Mechanism Associated with Leprosy Polarization. PLoS Pathog. 2017, 13, e1006103. [Google Scholar] [CrossRef] [PubMed]
- Scollard, D.; Adams, L.B.; Gillis, T.B.; Krahenbuhl, J.L.; Truman, R.W.; Williams, D.L. The continuing challenges of leprosy. Clin. Microbiol. Rev. 2006, 19, 338–381. [Google Scholar] [CrossRef] [Green Version]
- Tió-Coma, M.; van Hooij, A.; Bobosha, K.; Schip, J.J.P.; Banu, S.; Khadge, S.; Thapa, P.; Kunwar, C.B.; Goulart, I.M.; Bekele, Y.; et al. Whole Blood RNA Signatures in Leprosy Patients Identify Reversal Reactions Before Clinical Onset: A Prospective, Multicenter Study. Sci. Rep. 2019, 9, 17931. [Google Scholar] [CrossRef] [PubMed]
- Corstjens, P.L.; van Hooij, A.; Tjon Kon Fat, E.M.; van den Eeden, S.J.; Wilson, L.; Geluk, A. Field-friendly test for monitoring multiple immune response markers during onset and treatment of exacerbated immunity in leprosy. Clin. Vaccine Immunol. 2016, 23, 515–519. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Silva, C.A.; Webb, K.; Andre, B.G.; Marques, M.A.; Carvalho, F.M.; de Macedo, C.S.; Pinheiro, R.O.; Sarno, E.N.; Pessolani, M.C.; Belisle, J.T. Type 1 Reaction in Patients with Leprosy Corresponds to a Decrease in Proresolving Lipid Mediators and an Increase in Proinflammatory Lipid Mediators. J. Infect. Dis. 2017, 215, 431–439. [Google Scholar] [CrossRef] [PubMed]
- De Mattos Barbosa, M.G.; da Silva Prata, R.B.; Andrade, P.R.; Ferreira, H.; de Andrade Silva, B.J.; da Paixão de Oliveira, J.A.; Assis, T.Q.; de Toledo-Pinto, T.G.; de Lima Bezerra, O.C.; da Costa Nery, J.A.; et al. Indoleamine 2,3 dioxygenase and iron are required for Mycobacterium leprae survival. Microbes Infect. 2017, 11, 505–514. [Google Scholar] [CrossRef]
- Montoya, D.; Cruz, D.; Teles, R.M.B.; Lee, D.J.; Ochoa, M.T.; Krutzik, S.R.; Chun, R.; Schenk, M.; Zhang, X.; Ferguson, B.G.; et al. Divergence of macrophage phagocytic and antimicrobial programs in leprosy. Cell Host Microbe 2009, 6, 343–353. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Teles, R.M.B.; Graeber, T.G.; Krutzik, S.R.; Montoya, D.; Schenk, M.; Lee, D.J.; Komisopoulou, E.; Kelly-Scumpia, K.; Chun, R.; Iyer, S.S.; et al. Type I Interferon Suppresses Type II Interferon–Triggered Human Anti-Mycobacterial Responses. Science 2013, 339, 1448–1453. [Google Scholar] [CrossRef] [Green Version]
- Andrade, P.R.; Pinheiro, R.O.; Sales, A.M.; Illarramendi, X.; de Mattos Barbosa, M.G.; Moraes, M.O.; Jardim, M.R.; Nery, J.A.; Sampaio, E.P.; Sarno, E.N.; et al. Type 1 reaction in leprosy: A model for a better understanding of tissue immunity under an immunopathological condition. Expert Rev. Clin. Immunol. 2015, 11, 391–407. [Google Scholar] [CrossRef]
- Klionsky, D.J.; Abdel-Aziz, A.K.; Abdelfatah, S.; Abdellatif, M.; Abdoli, A.; Abel, S.; Abeliovich, H.; Abildgaard, M.H.; Abudu, Y.P.; Acevedo-Arozena, A.; et al. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy 2021, 17, 1–382. [Google Scholar] [CrossRef] [PubMed]
- De Mattos Barbosa, M.G.; de Andrade Silva, B.J.; Assis, T.Q.; da Silva Prata, R.B.; Ferreira, H.; Andrade, P.R.; da Paixão de Oliveira, J.A.; Sperandio da Silva, G.M.; da Costa Nery, J.A.; Sarno, E.M.; et al. Autophagy Impairment Is Associated with Increased Inflammasome Activation and Reversal Reaction Development in Multibacillary Leprosy. Front. Immunol. 2018, 9, 1223. [Google Scholar] [CrossRef]
- Ridley, D.S.; Jopling, W.H. Classification of leprosy according to immunity. A five-group system. Int. J. Lepr. Other Mycobact. Dis. 1966, 34, 255–273. [Google Scholar]
- Adams, L.B.; Soileau, N.A.; Battista, J.R.; Krahenbuhl, J.L. Inhibition of metabolism and growth of Mycobacterium leprae by gamma irradiation. Int. J. Lepr. Other Mycobact. Dis. 2000, 68, 1–10. [Google Scholar]
- Rovetta, A.I.; Peña, D.; Hernández Del Pino, R.E.; Recalde, G.M.; Pellegrini, J.; Bigi, F.; Musella, R.M.; Palmero, D.J.; Gutierrez, M.; Colombo, M.I.; et al. IFNG-mediated immune responses enhance autophagy against Mycobacterium tuberculosis antigens in patients with active tuberculosis. Autophagy 2014, 10, 2109–2121. [Google Scholar] [CrossRef] [Green Version]
- De Toledo-Pinto, T.G.; Ferreira, A.B.R.; Ribeiro-Alves, M.; Rodrigues, L.S.; Batista-Silva, L.R.; de Andrade Silva, B.J.; Lemes, R.M.; Martinez, A.N.; Sandoval, F.G.; Alvarado-Arnez, L.E.; et al. STING-Dependent 2’-5’ Oligoadenylate Synthetase-Like Production Is Required for Intracellular Mycobacterium leprae Survival. J. Infect. Dis. 2016, 214, 311–320. [Google Scholar] [CrossRef] [Green Version]
- Ramakers, C.; Ruijter, J.M.; Deprez, R.H.L.; Moorman, A.F.M. Assumption-free analysis of quantitative real-time PCR data. Neurosci. Lett. 2003, 339, 62–66. [Google Scholar] [CrossRef]
- Ruijter, J.M.; Ramakers, C.; Hoogaars, W.; Bakker, O.; van den Hoff, M.J.B.; Karlen, Y.; Moorman, A.F.M. Amplification efficiency: Linking baseline and bias in the analysis of quantitative PCR data. Nucleic Acids Res. 2009, 37, e45. [Google Scholar] [CrossRef] [Green Version]
- Schenk, M.; Krutzik, S.R.; Sieling, P.A.; Lee, D.J.; Teles, R.M.B.; Ochoa, M.T.; Komisopoulou, E.; Sarno, E.N.; Rea, T.H.; Graeber, T.G.; et al. NOD2 triggers an interleukin-32-dependent human dendritic cell program in leprosy. Nat. Med. 2012, 18, 555–563. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feng, Y.; He, D.; Yao, Z.; Klionsky, D.J. The machinery of macroautophagy. Cell Res. 2014, 24, 24–41. [Google Scholar] [CrossRef] [Green Version]
- Oliveros, J.C. VENNY. An. Interactive Tool for Comparing Lists with Venn Diagrams. 2007. Available online: https://bioinfogp.cnb.csic.es/tools/venny/index.html (accessed on 1 May 2021).
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef] [Green Version]
- Belone, A.d.F.F.; Rosa, P.S.; Trombone, A.P.F.; Fachin, L.R.V.; Guidella, C.C.; Ura, S.; Barreto, J.A.; Pinilla, M.G.; de Carvalho, A.F.; Carraro, D.M.; et al. Genome-wide screening of mRNA expression in leprosy patients. Front. Genet. 2015, 6, 334. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Münz, C. Enhancing immunity through autophagy. Annu. Rev. Immunol. 2009, 27, 423–449. [Google Scholar] [CrossRef]
- Travassos, L.H.; Carneiro, L.A.; Ramjeet, M.; Hussey, S.; Kim, Y.G.; Magalhães, J.G.; Yuan, L.; Soares, F.; Chea, E.; Le Bourhis, L.; et al. Nod1 and Nod2 direct autophagy by recruiting ATG16L1 to the plasma membrane at the site of bacterial entry. Nat. Immunol. 2010, 11, 55–62. [Google Scholar] [CrossRef] [PubMed]
- Khadge, S.; Banu, S.; Bobosha, K.; van Schip, J.J.v.d.P.; Goulart, I.M.; Thapa, P.; Kunwar, C.B.; van Meijgaarden, K.E.; van den Eeden, S.J.; Wilson, L.; et al. Longitudinal immune profiles in type 1 leprosy reactions in Bangladesh, Brazil, Ethiopia and Nepal. BMC Infect. Dis. 2015, 28, 15–477. [Google Scholar] [CrossRef] [Green Version]
- Walker, S.L.; Nicholls, P.P.; Butlin, C.R.; Nery, J.A.; Roy, H.K.; Rangel, E.; Sales, A.M.; Lockwood, D.N. Development and validation of a severity scale for leprosy type 1 reactions. PLoS Negl. Trop. Dis. 2008, 2, e351. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Geluk, A.; Bobosha, K.; van Schip, J.J.v.d.P.; Spencer, J.S.; Banu, S.; Martins, S.B.; Cho, S.N.; Franken, K.L.; Kim, H.J.; Bekele, Y.; et al. New biomarkers with relevance to leprosy diagnosis applicable in areas hyperendemic for leprosy. J. Immunol. 2012, 188, 4782–4791. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Geluk, A.; van Meijgaarden, K.E.; Wilson, L.; Bobosha, K.; van der Ploeg-van Schip, J.J.; van den Eeden, S.J.F.; Quinten, E.; Dijkman, K.; Franken, K.L.; Haisma, E.M.; et al. Longitudinal immune responses and gene expression profiles in type 1 leprosy reactions. J. Clin. Immunol. 2014, 34, 245–255. [Google Scholar] [CrossRef]
- Teles, R.M.B.; Lu, J.; Tió-Coma, M.; Goulart, I.M.B.; Banu, S.; Hagge, D.; Bobosha, K.; Ottenhoff, T.H.M.; Pellegrini, M.; Geluk, A.; et al. Identification of a systemic interferon-gamma inducible antimicrobial gene signature in leprosy patients undergoing reversal reaction. PLoS Negl. Trop. Dis. 2019, 13, e0007764. [Google Scholar] [CrossRef]
- Chaitanya, V.S.; Lavania, M.; Nigam, A.; Turankar, R.P.; Singh, I.; Horo, I.; Sengupta, U.; Jadhav, R.S. Cortisol and proinflammatory cytokine profiles in type 1 (reversal) reactions of leprosy. Immunol. Lett. 2013, 156, 159–167. [Google Scholar] [CrossRef]
- Lee, P.L.; Tang, Y.; Li, H.; Guertin, D.A. Raptor/mTORC1 loss in adipocytes causes progressive lipodystrophy and fatty liver disease. Mol. Metab. 2016, 5, 422–432. [Google Scholar] [CrossRef]
- Graef, M.; Friedman, J.R.; Graham, C.; Babu, M.; Nunnari, J. ER exit sites are physical and functional core autophagosome biogenesis components. Mol. Biol. Cell 2013, 24, 2918–2931. [Google Scholar] [CrossRef] [PubMed]
- Kirkin, V.; Lamark, T.; Johansen, T.; Dikic, I. NBR1 cooperates with p62 in selective autophagy of ubiquitinated targets. Autophagy 2009, 5, 732–733. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reichen, J.; Stickel, F.; Bhattacharya, I.; Matschke, K.; Maller, E.; Korth-Bradley, J. Repeat-dose sirolimus pharmacokinetics and pharmacodynamics in patients with hepatic allografts. Eur. J. Clin. Pharmacol. 2012, 68, 589–597. [Google Scholar] [CrossRef] [PubMed]
- Ge, Y.; Huang, M.; Yao, Y.M. Autophagy and proinflammatory cytokines: Interactions and clinical implications. Cytokine Growth Factor Rev. 2018, 43, 38–46. [Google Scholar] [CrossRef]
- Tokura, Y.; Phadungsaksawasdi, P.; Kurihara, K.; Fujiyama, T.; Honda, T. Pathophysiology of skin resident memory T cells. Front. Immunol. 2020, 11, 618897. [Google Scholar] [CrossRef]
- Dos Santos, L.N.; da Silva, P.H.; Alvim, I.M.; Nery, J.A.; Lara, F.A.; Sarno, E.N.; Esquenazi, D. Role of Teffector/memory cells, TBX1 gene expression and T-cell homing receptor on type 1 reaction in borderline lepromatous leprosy patients. PLoS ONE 2016, 11, e0164543. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheuk, S.; Schlums, H.; Gallais Sérézal, I.; Martini, E.; Chiang, S.C.; Marquardt, N.; Gibbs, A.; Detlofsson, E.; Introini, A.; Forkel, M.; et al. CD49a Expression Defines Tissue-Resident CD8+T Cells Poised for Cytotoxic Function in Human Skin. Immunity 2017, 46, 287–300. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Oliveira, A.L.; Amadeu, T.P.; de França Gomes, A.C.; Menezes, V.M.; da Costa Nery, J.A.; Pinheiro, R.O.; Sarno, E.N. Role of CD8(+) T cells in triggering reversal reaction in HIV/leprosy patients. Immunology 2013, 140, 47–60. [Google Scholar] [CrossRef]
- Fabri, M.; Stenger, S.; Shin, D.M.; Yuk, J.M.; Liu, P.T.; Realegeno, S.; Lee, H.M.; Krutzik, S.R.; Schenk, M.; Sieling, P.A.; et al. Vitamin D is required for IFNgamma- mediated antimicrobial activity of human macrophages. Sci. Transl. Med. 2011, 3, 104ra102. [Google Scholar] [CrossRef] [Green Version]
- Yuk, J.M.; Shin, D.M.; Lee, H.M.; Yang, C.S.; Jin, H.S.; Kim, K.K.; Lee, Z.W.; Lee, S.H.; Kim, J.M.; Jo, E.K. Vitamin D3 induces autophagy in human monocytes/macrophages via cathelicidin. Cell Host Microbe 2009, 6, 231–243. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Lavender, P.; Watson, J.; Arno, M.; Lehner, T. Stress-activated Dendritic Cells (DC) Induce Dual Interleukin (IL)-15- and IL1_-mediated Pathways, Which May Elicit CD4_ Memory T Cells and Interferon (IFN)-stimulated Genes. J. Biol. Chem. 2015, 290, 15595–15609. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cooper, C.L.; Mueller, C.; Sinchaisri, T.A.; Pirmez, C.; Chan, J.; Kaplan, G.; Young, S.M.; Weissman, I.L.; Bloom, B.R.; Rea, T.H.; et al. Analysis of naturally occurring delayed-type hypersensitivity reactions in leprosy by in situ hybridization. J. Exp. Med. 1989, 169, 1565–1581. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Richter, S.; Geldner, N.; Schrader, J.; Wolters, H.; Stierhof, Y.D.; Rios, G.; Koncz, C.; Robinson, D.G.; Jürgens, G. Functional diversification of closely related ARF-GEFs in protein secretion and recycling. Nature 2007, 448, 488–492. [Google Scholar] [CrossRef]
- Zanetti, G.; Pahuja, K.B.; Studer, S.; Shim, S.; Schekman, R. COPII and the regulation of protein sorting in mammals. Nat. Cell Biol. 2011, 14, 20–28. [Google Scholar] [CrossRef]
- Amodio, G.; Venditti, R.; De Matteis, M.A.; Moltedo, O.; Pignataro, P.; Remondelli, P. Endoplasmic reticulum stress reduces COPII vesicle formation and modifies Sec23a cycling at ERESs. FEBS Lett. 2013, 587, 3261–3266. [Google Scholar] [CrossRef] [Green Version]
- Jing, J.; Wang, B.; Liu, P. The Functional Role of SEC23 in Vesicle Transportation, Autophagy and Cancer. Int. J. Biol. Sci. 2019, 15, 2419–2426. [Google Scholar] [CrossRef] [Green Version]
- Ishihara, N.; Hamasaki, M.; Yokota, S.; Suzuki, K.; Kamada, Y.; Kihara, A.; Yoshimori, T.; Noda, T.; Ohsumi, Y. Autophagosome requires specific early Sec proteins for its formation and NSF/SNARE for vacuolar fusion. Mol. Biol. Cell 2001, 12, 3690–3702. [Google Scholar] [CrossRef]
- Reggiori, F.; Wang, C.W.; Nair, U.; Shintani, T.; Abeliovich, H.; Klionsky, D.J. Early stages of the secretory pathway, but not endosomes, are required for Cvt vesicle and autophagosome assembly in Saccharomyces cerevisiae. Mol. Biol. Cell 2004, 15, 2189–2204. [Google Scholar] [CrossRef] [Green Version]
- Lemus, L.; Ribas, J.L.; Sikorska, N.; Goder, V. An ER-Localized SNARE Protein Is Exported in Specific COPII Vesicles for Autophagosome Biogenesis. Cell Rep. 2016, 14, 1710–1722. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tan, D.; Cai, Y.; Wang, J.; Zhang, J.; Menon, S.; Chou, H.-T.; Ferro-Novick, S.; Reinisch, K.M.; Walz, T. The EM structure of the TRAPPIII complex leads to the identification of a requirement for COPII vesicles on the macroautophagy pathway. Proc. Natl. Acad. Sci. USA 2013, 110, 19432–19437. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dewi, F.R.P.; Jiapaer, S.; Kobayashi, A.; Hazawa, M.; Ikliptikawati, D.K.; Hartono; Sabit, H.; Nakada, M.; Wong, R.W. Nucleoporin TPR (translocated promoter region, nuclear basket protein) upregulation alters MTOR-HSF1 trails and suppresses autophagy induction in ependymoma. Autophagy 2020, 24, 1–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Funasaka, T.; Tsuka, E.; Wong, R.W. Regulation of autophagy by nucleoporin Tpr. Sci. Rep. 2012, 2, 878. [Google Scholar] [CrossRef] [Green Version]
- Mochizuki, N.; Cho, G.; Wen, B.; Insel, P.A. Identification and cDNA cloning of a novel human mosaic protein, LGN, based on interaction with G alpha i2. Gene 1996, 181, 39–43. [Google Scholar] [CrossRef]







| PB | MB No Progression | MB Progression | T1R | |
|---|---|---|---|---|
| Male/female | 6/8 | 7/1 | 5/2 | 9/3 |
| Age, mean (range) | 54.5 (8–92) | 53.37 (34–65) | 45.14 (32–69) | 49.16 (17–66) |
| BI, mean (range) | 0 (0–0) | 2.15 (1.50–5.50) | 2.98 (0.50–4.67) | 2.66 (0.75–5.85) |
| LBI (range) | 0 (0–0) | 4.38 (2.85–5.95) | 4.6 (2.7–5.95) | 2.06 (0–3.8) |
| Ridley–Jopling Clinical Form of Leprosy, n | ||||
| BT | 14 | - | - | - |
| BL | - | 4 | 4 | 9 |
| LL | - | 4 | 3 | 3 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Silva, B.J.d.A.; Bittencourt, T.L.; Leal-Calvo, T.; Mendes, M.A.; Prata, R.B.d.S.; Barbosa, M.G.d.M.; Andrade, P.R.; Côrte-Real, S.; Sperandio da Silva, G.M.; Moraes, M.O.; et al. Autophagy-Associated IL-15 Production Is Involved in the Pathogenesis of Leprosy Type 1 Reaction. Cells 2021, 10, 2215. https://doi.org/10.3390/cells10092215
Silva BJdA, Bittencourt TL, Leal-Calvo T, Mendes MA, Prata RBdS, Barbosa MGdM, Andrade PR, Côrte-Real S, Sperandio da Silva GM, Moraes MO, et al. Autophagy-Associated IL-15 Production Is Involved in the Pathogenesis of Leprosy Type 1 Reaction. Cells. 2021; 10(9):2215. https://doi.org/10.3390/cells10092215
Chicago/Turabian StyleSilva, Bruno Jorge de Andrade, Tamiris Lameira Bittencourt, Thyago Leal-Calvo, Mayara Abud Mendes, Rhana Berto da Silva Prata, Mayara Garcia de Mattos Barbosa, Priscila Ribeiro Andrade, Suzana Côrte-Real, Gilberto Marcelo Sperandio da Silva, Milton Ozório Moraes, and et al. 2021. "Autophagy-Associated IL-15 Production Is Involved in the Pathogenesis of Leprosy Type 1 Reaction" Cells 10, no. 9: 2215. https://doi.org/10.3390/cells10092215
APA StyleSilva, B. J. d. A., Bittencourt, T. L., Leal-Calvo, T., Mendes, M. A., Prata, R. B. d. S., Barbosa, M. G. d. M., Andrade, P. R., Côrte-Real, S., Sperandio da Silva, G. M., Moraes, M. O., Sarno, E. N., & Pinheiro, R. O. (2021). Autophagy-Associated IL-15 Production Is Involved in the Pathogenesis of Leprosy Type 1 Reaction. Cells, 10(9), 2215. https://doi.org/10.3390/cells10092215