In Vitro Differentiation of Human Amniotic Epithelial Cells into Hepatocyte-like Cells
Abstract
:1. Introduction
2. Materials and Methods
2.1. Human Placenta
2.2. Isolation of Amniotic Epithelial Cells
2.3. Cell Culture
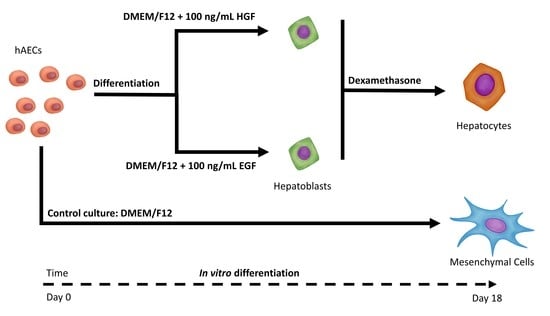
2.3.1. hAEC Differentiation
2.3.2. Collection of Cells for Research and Passage
2.4. Cytometric Analysis of Surface Markers
2.5. Cell Count and Viability Assessment
2.6. Immunofluorescent Identification of Selected Proteins
2.7. Assessment of the Ability of Cells to Secrete Albumin
2.8. Assessment of the Ability of Cells to Synthesise and Store Glycogen
2.9. Real-Time Quantitative Polymerase Chain Reaction (RT-qPCR)
2.10. Statistical Analysis
3. Results
3.1. Cell Morphology
3.2. Cell Count and Viability Assessment
3.3. Analysis of Expression and Co-Expression of Mesenchymal Surface Markers and the SSEA-4 Pluripotent Cell Marker
3.4. Gene Expression Analysis of Pluripotency and Germ Layer Markers
3.5. Expression Analysis of mRNA and Protein Markers of the Hepatoblast/Hepatocyte Lineage
3.6. Functional Assessment of Differentiated Cells
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Weiler, N.; Schlotmann, A.; Schnitzbauer, A.A.; Zeuzem, S.; Welker, M.W. The Epidemiology of Acute Liver Failure: Results of a Population-Based Study Including 25 Million State-Insured Individuals. Dtsch. Ärztebl. Int. 2020, 117, 43–50. [Google Scholar] [CrossRef] [PubMed]
- Bernal, W.; Wendon, J. Acute Liver Failure. N. Engl. J. Med. 2013, 369, 2525–2559. [Google Scholar] [CrossRef]
- Thorburn, D.; Taylor, R.; Whitney, J.; Adair, A.; Attia, M.; Gibbs, P.; Grammatikopoulos, T.; Isaac, R.J.; Masson, S.; Marshall, A.; et al. Resuming Liver Transplantation amid the COVID-19 Pandemic. Lancet 2021, 6, 12–13. [Google Scholar] [CrossRef]
- Grande, R.G.; Pérez, M.J. Liver Transplantation in Acute Liver Failure: Indications and Outcome. In Liver Research and Clinical Management; Rodrigo, A., Ed.; InTechOpen: London, UK, 2018; pp. 157–166. [Google Scholar] [CrossRef] [Green Version]
- Shi, Y.; Inoue, H.; Wu, J.C.; Yamanaka, S. Induced Pluripotent Stem Cell Technology: A Decade of Progress. Nat. Rev. Drug Discov. 2017, 16, 115–130. [Google Scholar] [CrossRef] [PubMed]
- Golchin, A.; Chatziparasidou, A.; Ranjbarvan, P.; Niknam, Z.; Ardeshirylajimi, A. Embryonic Stem Cells in Clinical Trials: Current Overview of Developments and Challenges. Adv. Exp. Med. Biol. 2021, 1312, 19–37. [Google Scholar] [CrossRef]
- al Abbar, A.; Ngai, S.C.; Nograles, N.; Alhaji, S.Y.; Abdullah, S. Induced Pluripotent Stem Cells: Reprogramming Platforms and Applications in Cell Replacement Therapy. BioRes. Open Access 2020, 9, 121–136. [Google Scholar] [CrossRef]
- Yasuda, S.Y.; Ikeda, T.; Shahsavarani, H.; Yoshida, N.; Nayer, B.; Hino, M.; Vartak-Sharma, N.; Suemori, H.; Hasegawa, K. Chemically Defined and Growth-Factor-Free Culture System for the Expansion and Derivation of Human Pluripotent Stem Cells. Nat. Biomed. Eng. 2018, 2, 173–182. [Google Scholar] [CrossRef]
- Doss, M.X.; Sachinidis, A. Current Challenges of IPSC-Based Disease Modeling and Therapeutic Implications. Cells 2019, 8, 403. [Google Scholar] [CrossRef] [Green Version]
- Gorecka, J.; Kostiuk, V.; Fereydooni, A.; Gonzalez, L.; Luo, J.; Dash, B.; Isaji, T.; Ono, S.; Liu, S.; Lee, S.R.; et al. The Potential and Limitations of Induced Pluripotent Stem Cells to Achieve Wound Healing. Stem Cell Res. Ther. 2019, 10, 87. [Google Scholar] [CrossRef] [Green Version]
- Szkolnicka, D.; Farnworth, S.L.; Lucendo-Villarin, B.; Storck, C.; Zhou, W.; Iredale, J.P.; Flint, O.; Hay, D.C. Accurate Prediction of Drug-Induced Liver Injury Using Stem Cell-Derived Populations. Stem Cells Transl. Med. 2014, 3, 141–148. [Google Scholar] [CrossRef]
- Basma, H.; Soto-Gutiérrez, A.; Yannam, G.R.; Liu, L.; Ito, R.; Yamamoto, T.; Ellis, E.; Carson, S.D.; Sato, S.; Chen, Y.; et al. Differentiation and Transplantation of Human Embryonic Stem Cell-Derived Hepatocytes. Gastroenterology 2009, 136, 990–999. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Si-Tayeb, K.; Noto, F.K.; Nagaoka, M.; Li, J.; Battle, M.A.; Duris, C.; North, P.E.; Dalton, S.; Duncan, S.A. Highly Efficient Generation of Human Hepatocyte-like Cells from Induced Pluripotent Stem Cells. Hepatology 2010, 51, 297–305. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Farzaneh, Z.; Pakzad, M.; Vosough, M.; Pournasr, B.; Baharvand, H. Differentiation of Human Embryonic Stem Cells to Hepatocyte-like Cells on a New Developed Xeno-Free Extracellular Matrix. Histochem. Cell Biol. 2014, 142, 217–226. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.; Vizetto-Duarte, C.; Moay, Z.K.; Setyawati, M.I.; Rakshit, M.; Kathawala, M.H.; Ng, K.W. Composite Hydrogels in Three-Dimensional in Vitro Models. Front. Bioeng. Biotechnol. 2020, 8, 611. [Google Scholar] [CrossRef]
- Carpentier, A.; Nimgaonkar, I.; Chu, V.; Xia, Y.; Hu, Z.; Liang, T.J. Hepatic Differentiation of Human Pluripotent Stem Cells in Miniaturized Format Suitable for High-Throughput Screen. Stem Cell Res. 2016, 16, 640–650. [Google Scholar] [CrossRef] [Green Version]
- Gramignoli, R.; Srinivasan, R.C.; Kannisto, K.; Strom, S.C. Isolation of Human Amnion Epithelial Cells According to Current Good Manufacturing Procedures. Curr. Protoc. Stem Cell Biol. 2016, 37, 1E.10.1–1E.10.13. [Google Scholar] [CrossRef]
- Szmytkowska, P.; Koryciak-Komarska, H.; Limanówka, Ł.; Król, M.; Plewka, D.; Kopaczka, K.; Czekaj, P. Comparison on Efficiency of Various Enzymatic methods for the Isolation of Cells from Human Amnion. In Advances in Biomedical Research. From Microbiology to Cancer; Biały, Ł., Młynarczuk-Biały, M., Eds.; Wydawnictwo Naukowe Tygiel Lublin-Warszawa: Lublin, Polska, 2019; pp. 99–110. [Google Scholar]
- Gramignoli, R.; Morandi, F.; Horenstein, A.; Strunz, B.; Srinivasan, R.C.; Malavasi, F.; Strom, S.C. Cellular Mechanism in Support of Allogenic Human Amnion Epithelial Cell Transplantation without Immunosuppression. Cythotherapy 2019, 21, S26. [Google Scholar] [CrossRef]
- Kuk, N.; Hodge, A.; Sun, Y.; Correia, J.; Alhomrani, M.; Samuel, C.; Moore, G.; Lim, R.; Sievert, W. Human Amnion Epithelial Cells and Their Soluble Factors Reduce Liver Fibrosis in Murine Non-Alcoholic Steatohepatitis. J. Gastroenterol. Hepatol. 2019, 34, 1441–1449. [Google Scholar] [CrossRef]
- Cargnoni, A.; Farigu, S.; Cotti Piccinelli, E.; Bonassi Signoroni, P.; Romele, P.; Vanosi, G.; Toschi, I.; Cesari, V.; Barros Sant’Anna, L.; Magatti, M.; et al. Effect of Human Amniotic Epithelial Cells on Pro-Fibrogenic Resident Hepatic Cells in a Rat Model of Liver Fibrosis. J. Cell Mol. Med. 2018, 22, 1202–1213. [Google Scholar] [CrossRef] [Green Version]
- Rodriguez, N.S.; Yanuaria, L.; Parducho, K.M.R.; Garcia, I.M.; Varghese, B.A.; Grubbs, B.H.; Miki, T. Liver-Directed Human Amniotic Epithelial Cell Transplantation Improves Systemic Disease Phenotype in Hurler Syndrome Mouse Model. Stem Cells Transl. Med. 2017, 6, 1583–1594. [Google Scholar] [CrossRef] [Green Version]
- Czekaj, P.; Król, M.; Limanówka, Ł.; Michalik, M.; Lorek, K.; Gramignoli, R. Assessment of Animal Experimental Models of Toxic Liver Injury in the Context of Their Potential Application as Preclinical Models for Cell Therapy. Eur. J. Pharmacol. 2019, 861, 172597. [Google Scholar] [CrossRef] [PubMed]
- Alhomrani, M.; Correia, J.; Zavou, M.; Leaw, B.; Kuk, N.; Xu, R.; Saad, M.I.; Hodge, A.; Greening, D.W.; Lim, R.; et al. The Human Amnion Epithelial Cell Secretome Decreases Hepatic Fibrosis in Mice with Chronic Liver Fibrosis. Front. Pharmacol. 2017, 8, 748. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.W.; Liu, Q.Y.; Li, J.Y.; Wei, L.; Ren, K.K.; Zhang, X.C.; Ding, T.; Xiao, L.; Zhang, W.J.; Wu, H.Y.; et al. Therapeutic Efficiency of Human Amniotic Epithelial Stem Cell-Derived Functional Hepatocyte-like Cells in Mice with Acute Hepatic Failure. Stem Cell Res. Ther. 2018, 9, 321. [Google Scholar] [CrossRef] [PubMed]
- Strom, S.C.; Gramignoli, R. Human Amnion Epithelial Cells Expressing HLA-G as Novel Cell-Based Treatment for Liver Disease. Hum. Immunol. 2016, 77, 734–739. [Google Scholar] [CrossRef] [PubMed]
- Michalik, M.; Gładyś, A.; Czekaj, P. Differentiation of Cells Isolated from Afterbirth Tissues into Hepatocyte-Like Cells and Their Potential Clinical Application in Liver Regeneration. Stem Cell Rev. Rep. 2021, 17, 581–603. [Google Scholar] [CrossRef]
- Fanti, M.; Gramignoli, R.; Serra, M.; Cadoni, E.; Strom, C.S.; Marongiu, F. Differentiation of amniotic epithelial cells into various liver cell types and potential therapeutic applications. Placenta 2017, 59, 139–145. [Google Scholar] [CrossRef]
- Marongiu, F.; Gramignoli, R.; Dorko, K.; Miki, T.; Ranade, A.R.; Serra, M.P.; Doratiotto, S.; Sini, M.; Sharma, S.; Mitamura, K.; et al. Hepatic Differentiation of Amniotic Epithelial Cells. Hepatology 2011, 53, 1719–1729. [Google Scholar] [CrossRef] [Green Version]
- Coronado, E.R.; Somaraki-Cormier, M.; Ong, L.J.; Halff, A.G. Hepatocyte-like cells derived from human amniotic epithelial, bone marrow, and adipose stromal cells display enhanced functionality when cultured on decellularized liver substrate. Stem Cell Res. 2019, 38, 101471. [Google Scholar] [CrossRef]
- Bilic, G.; Zeisberger, S.M.; Mallik, A.S.; Zimmermann, R.; Zisch, A.H. Comparative Characterization of Cultured Human Term Amnion Epithelial and Mesenchymal Stromal Cells for Application in Cell Therapy. Cell Transpl. 2008, 17, 955–968. [Google Scholar] [CrossRef] [Green Version]
- Lamouille, S.; Xu, J.; Derynck, R. Molecular Mechanisms of Epithelial-Mesenchymal Transition. Nat. Rev. Mol. Cell Biol. 2014, 15, 178–196. [Google Scholar] [CrossRef] [Green Version]
- Lin, J.S.; Zhou, L.; Sagayaraj, A.; Jumat, N.H.B.; Choolani, M.; Chan, J.K.Y.; Biswas, A.; Wong, P.C.; Lim, S.G.; Dan, Y.Y. Hepatic Differentiation of Human Amniotic Epithelial Cells and in Vivo Therapeutic Effect on Animal Model of Cirrhosis. J. Gastroenterol. Hepatol. 2015, 30, 1673–1682. [Google Scholar] [CrossRef] [PubMed]
- Miki, T.; Marongiu, F.; Ellis, E.C.S.; Dorko, K.; Mitamura, K.; Ranade, A.; Gramignoli, R.; Davila, J.; Strom, S.C. Production of Hepatocyte-like Cells from Human Amnion. Methods Mol. Biol. 2009, 481, 155–168. [Google Scholar] [CrossRef] [PubMed]
- Xie, F.; Xiao, P.; Chen, D.; Xu, L.; Zhang, B. MiRDeepFinder: A MiRNA Analysis Tool for Deep Sequencing of Plant Small RNAs. Plant Mol. Biol. 2012, 80, 75–84. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Lai, D. Application of Human Amniotic Epithelial Cells in Regenerative Medicine: A Systematic Review. Stem Cell Res. Ther. 2020, 11, 439. [Google Scholar] [CrossRef]
- Liu, J.; Wen, Y.; Luo, W.; Liu, Y.; Sha, X. Human Amniotic Epithelial Cells Promote the Proliferation of Human Corneal Endothelial Cells by Regulating Telomerase Activity via the Wnt/β-Catenin Pathway. Curr. Eye Res. 2020, 46, 159–167. [Google Scholar] [CrossRef]
- Maymó, J.L.; Riedel, R.; Pérez-Pérez, A.; Magatti, M.; Maskin, B.; Dueñas, J.L.; Parolini, O.; Sánchez-Margalet, V.; Varone, C.L. Proliferation and Survival of Human Amniotic Epithelial Cells during Their Hepatic Differentiation. PLoS ONE 2018, 13, e0191489. [Google Scholar] [CrossRef] [Green Version]
- Yang, P.-J.; Yuan, W.-X.; Liu, J.; Li, J.-Y.; Tan, B.; Qiu, C.; Zhu, X.-L.; Qiu, C.; Lai, D.-M.; Guo, L.-H.; et al. Biological Characterization of Human Amniotic Epithelial Cells in a Serum-Free System and Their Safety Evaluation. Acta Pharmacol. Sin. 2018, 39, 1305–1316. [Google Scholar] [CrossRef]
- Bejaoui, M.; Ferdousi, F.; Zheng, Y.W.; Oda, T.; Isoda, H. Regulating Cell Fate of Human Amnion Epithelial Cells Using Natural Compounds: An Example of Enhanced Neural and Pigment Differentiation by 3,4,5-Tri-O-Caffeoylquinic Acid. Cell Commun. Signal. 2021, 19, 26. [Google Scholar] [CrossRef]
- Kolanko, E.; Kopaczka, K.; Koryciak-Komarska, H.; Czech, E.; Szmytkowska, P.; Gramignoli, R.; Czekaj, P. Increased Immunomodulatory Capacity of Human Amniotic Cells after Activation by Pro-Inflammatory Chemokines. Eur. J. Pharmacol. 2019, 859, 172545. [Google Scholar] [CrossRef]
- Jaramillo, M.; Yeh, H.; Yarmush, M.L.; Uygun, B.E. Decellularized Human Liver Extracellular Matrix (HDLM)-Mediated Hepatic Differentiation of Human Induced Pluripotent Stem Cells (HIPSCs). J. Tissue Eng. Regen. Med. 2018, 12, 1962–1973. [Google Scholar] [CrossRef]
- Passaretta, F.; Bosco, D.; Centurione, L.; Centurione, M.A.; Marongiu, F.; di Pietro, R. Differential Response to Hepatic Differentiation Stimuli of Amniotic Epithelial Cells Isolated from Four Regions of the Amniotic Membrane. J. Cell Mol. Med. 2020, 24, 4350–4355. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Page, J.L.; Johnson, M.C.; Olsavsky, K.M.; Strom, S.C.; Zarbl, H.; Omiecinski, C.I. Gene Expression Profiling of Extracellular Matrix as an Effector of Human Hepatocyte Phenotype in Primary Cell Culture. Toxicol. Sci. 2007, 97, 384–397. [Google Scholar] [CrossRef] [PubMed]
- Sullivan, G.J.; Hay, D.C.; Park, I.H.; Fletcher, J.; Hannoun, Z.; Payne, C.M.; Dalgetty, D.; Black, J.R.; Ross, J.A.; Samuel, K.; et al. Generation of Functional Human Hepatic Endoderm from Human Induced Pluripotent Stem Cells. Hepatology 2010, 51, 329–335. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gorfien, S.F.; Jayme, D.W. Development and Optimization of Serum- and Protein-Free Culture Media. In Animal Cell Culture: Essential Methods; Davis, J.M., Ed.; Wiley-Blackwell: Chichester, UK, 2011; pp. 153–184. [Google Scholar]
- Miki, T.; Lehmann, T.; Cai, H.; Stolz, D.B.; Strom, S.C. Stem Cell Characteristics of Amniotic Epithelial Cells. Stem Cells 2005, 23, 1549–1559. [Google Scholar] [CrossRef] [Green Version]
- Tabatabaei, M.; Mosaffa, N.; Nikoo, S.; Bozorgmehr, M.; Ghods, R.; Kazemnejad, S.; Rezania, S.; Keshavarzi, B.; Arefi, S.; Ramezani-Tehrani, F.; et al. Isolation and Partial Characterization of Human Amniotic Epithelial Cells: The Effect of Trypsin. Avicenna J. Med. Biotechnol. 2014, 6, 10–20. [Google Scholar]
- Manuelpillai, U.; Moodley, Y.; Borlongan, C.V.; Parolini, O. Amniotic Membrane and Amniotic Cells: Potential Therapeutic Tools to Combat Tissue Inflammation and Fibrosis? Placenta 2011, 32, S320–S325. [Google Scholar] [CrossRef]
- Motedayyen, H.; Esmaeil, N.; Tajik, N.; Khadem, F.; Ghotloo, S.; Khani, B.; Rezaei, A. Method and Key Points for Isolation of Human Amniotic Epithelial Cells with High Yield, Viability and Purity. BMC Res. Notes 2017, 10, 552. [Google Scholar] [CrossRef] [Green Version]
- Murphy, S.; Rosli, S.; Acharya, R.; Mathias, L.; Lim, R.; Wallace, E.; Jenkin, G. Amnion Epithelial Cell Isolation and Characterization for Clinical Use. Curr. Protoc. Stem Cell Biol. 2010, 13, 1E.6.1–1E.6.25. [Google Scholar] [CrossRef]
- Murphy, S.V.; Kidyoor, A.; Reid, T.; Atala, A.; Wallace, E.M.; Lim, R. Isolation, Cryopreservation and Culture of Human Amnion Epithelial Cells for Clinical Applications. J. Vis. Exp. 2014, 94, e52085. [Google Scholar] [CrossRef] [Green Version]
- Srinivasan, R.C.; Strom, S.C.; Gramignoli, R. Effects of Cryogenic Storage on Human Amnion Epithelial Cells. Cells 2020, 9, 1696. [Google Scholar] [CrossRef]
- Pratama, G.; Vaghjiani, V.; Tee, J.Y.; Liu, Y.H.; Chan, J.; Tan, C.; Murthi, P.; Gargett, C.; Manuelpillai, U. Changes in Culture Expanded Human Amniotic Epithelial Cells: Implications for Potential Therapeutic Applications. PLoS ONE 2011, 6, e26136. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miki, T. Stem Cell Characteristics and the Therapeutic Potential of Amniotic Epithelial Cells. Am. J. Reprod. Immunol. 2018, 80, e13003. [Google Scholar] [CrossRef] [PubMed]
- Uchida, Y.; Ferdousi, F.; Zheng, Y.W.; Oda, T.; Isoda, H. Global Gene Expression Profiling Reveals Isorhamnetin Induces Hepatic-Lineage Specific Differentiation in Human Amniotic Epithelial Cells. Front. Cell Dev. Biol. 2020, 8, 578036. [Google Scholar] [CrossRef] [PubMed]
- Takashima, S.; Ise, H.; Zhao, P.; Akaike, T.; Nikaido, T. Human Amniotic Epithelial Cells Possess Hepatocyte-like Characteristics and Functions. Cell Struct. Funct. 2004, 29, 73–84. [Google Scholar] [CrossRef] [Green Version]
- Gomez Dominguez, R. Amniotische Epithelzellen Isolierung und Charakterisierung = Human Amniotic Epithelial Cells; VVB Laufersweiler: Giessen, Germany, 2008. [Google Scholar]
- Weidinger, A.; Poženel, L.; Wolbank, S.; Banerjee, A. Sub-Regional Differences of the Human Amniotic Membrane and Their Potential Impact on Tissue Regeneration Application. Front. Bioeng. Biotechnol. 2021, 8, 613804. [Google Scholar] [CrossRef] [PubMed]
- Söderdahl, T.; Küppers-Munther, B.; Heins, N.; Edsbagge, J.; Björquist, P.; Cotgreave, I.; Jernström, B. Glutathione Transferases in Hepatocyte-like Cells Derived from Human Embryonic Stem Cells. Toxicol. In Vitro 2007, 21, 929–937. [Google Scholar] [CrossRef]
- Mathew, J.; Cattan, A.R.; Hall, A.G.; Hines, J.E.; Nelson, R.; Eastham, E.; Burt, A.D. Glutathione S-Transferases in Neonatal Liver Disease. J. Clin. Pathol. 1992, 45, 679–683. [Google Scholar] [CrossRef] [Green Version]
- Jochum, C.; Beste, M.; Sowa, J.-P.; Farahani, M.; Penndorf, V.; Nadalin, S.; Saner, F.; Canbay, A.; Gerken, G. Glutathione-S-Transferase Subtypes α and π as a Tool to Predict and Monitor Graft Failure or Regeneration in a Pilot Study of Living Donor Liver Transplantation. Eur. J. Med. Res. 2011, 16, 34–40. [Google Scholar] [CrossRef] [Green Version]
- di Lollo, V.; Canciello, A.; Orsini, M.; Bernabò, N.; Ancora, M.; di Federico, M.; Curini, V.; Mattioli, M.; Russo, V.; Mauro, A.; et al. Transcriptomic and Computational Analysis Identified LPA Metabolism, KLHL14 and KCNE3 as Novel Regulators of Epithelial-Mesenchymal Transition. Sci. Rep. 2020, 10, 4180. [Google Scholar] [CrossRef]
- Alcaraz, A.; Mrowiec, A.; Insausti, C.L.; García-Vizcaíno, E.M.; Ruiz-Canada, C.; López-Martínez, M.C.; Moraleda, J.M.; Nicolás, F.J. Autocrine TGF-β Induces Epithelial to Mesenchymal Transition in Human Amniotic Epithelial Cells. Cell Trans. 2013, 22, 1351–1367. [Google Scholar] [CrossRef]
- Vallier, L.; Touboul, T.; Chng, Z.; Brimpari, M.; Hannan, N.; Millan, E.; Smithers, L.E.; Trotter, M.; Rugg-Gunn, P.; Weber, A.; et al. Early Cell Fate Decisions of Human Embryonic Stem Cells and Mouse Epiblast Stem Cells Are Controlled by the Same Signalling Pathways. PLoS ONE 2009, 4, e6082. [Google Scholar] [CrossRef]
- Matsukuma, S.; Takeo, H.; Kono, T.; Nagata, Y.; Sato, K. Aberrant Cytokeratin 7 Expression of Centrilobular Hepatocytes: A Clinicopathological Study. Histopathology 2012, 61, 857–862. [Google Scholar] [CrossRef] [PubMed]
- Paku, S.; Dezso, K.; Kopper, L.; Nagy, P. Immunohistochemical Analysis of Cytokeratin 7 Expression in Resting and Proliferating Biliary Structures of Rat Liver. Hepatology 2005, 42, 863–870. [Google Scholar] [CrossRef] [PubMed]
- Yan, B.; Jiang, Z.; Cheng, L.; Chen, K.; Zhou, C.; Sun, L.; Qian, W.; Li, J.; Cao, J.; Xu, Q.; et al. Paracrine HGF/c-MET Enhances the Stem Cell-like Potential and Glycolysis of Pancreatic Cancer Cells via Activation of YAP/HIF-1α. Exp. Cell Res. 2018, 371, 63–71. [Google Scholar] [CrossRef] [PubMed]
- Cao, Z.; Xie, Y.; Yu, L.; Li, Y. Hepatocyte Growth Factor (HGF) and Stem Cell Factor (SCF) Maintained the Stemness of Human Bone Marrow Mesenchymal Stem Cells (HBMSCs) during Long-Term Expansion by Preserving Mitochondrial Function via the PI3K/AKT, ERK1/2, and STAT3 Signaling Pathways. Stem Cell Res. Ther. 2020, 11, 329. [Google Scholar] [CrossRef]
- Yamasaki, C.; Tateno, C.; Aratani, A.; Ohnishi, C.; Katayama, S.; Kohashi, T.; Hino, H.; Marusawa, H.; Asahara, T.; Yoshizato, K. Growth and Differentiation of Colony-Forming Human Hepatocytes in Vitro. J. Hepatol. 2006, 44, 749–757. [Google Scholar] [CrossRef] [Green Version]
- Bogacheva, M.S.; Khan, S.; Kanninen, L.K.; Yliperttula, M.; Leung, A.W.; Lou, Y.R. Differences in Definitive Endoderm Induction Approaches Using Growth Factors and Small Molecules. J. Cell Physiol. 2018, 233, 3578–3589. [Google Scholar] [CrossRef]
- Chiang, J.Y.L.; Ferrell, J.M. Up to Date on Cholesterol 7 Alpha-Hydroxylase (CYP7A1) in Bile Acid Synthesis. Liver Res. 2020, 4, 47–63. [Google Scholar] [CrossRef]
- Li, T.; Chanda, D.; Zhang, Y.; SikChoi, H.; Chiang, J.Y.L. Glucose Stimulates Cholesterol 7 α-Hydroxylase Gene Transcription in Human Hepatocytes. J. Lipid. Res. 2010, 51, 832–842. [Google Scholar] [CrossRef] [Green Version]
- Vaghjiani, V.; Vaithilingam, V.; Saraswati, I.; Sali, A.; Murthi, P.; Kalionis, B.; Tuch, B.E.; Manuelpillai, U. Hepatocyte-like Cells Derived from Human Amniotic Epithelial Cells Can Be Encapsulated without Loss of Viability or Function in Vitro. Stem Cells Dev. 2014, 23, 866–876. [Google Scholar] [CrossRef] [Green Version]
- Shedge, S.; Roy, P.; Shedge, A.; Doshi, M.A.; Roy, P.P. Periodic Acid Schiff (PAS) Staining: A Useful Technique for Demonstration of Carbohydrates. Med. Legal Update 2020, 20, 353–357. [Google Scholar] [CrossRef]
- Elphick, L.M.; Pawolleck, N.; Guschina, I.A.; Chaieb, L.; Eikel, D.; Nau, H.; Harwood, J.L.; Plant, N.J.; Williams, R.S.B. Conserved Valproic-Acid-Induced Lipid Droplet Formation in Dictyostelium and Human Hepatocytes Identifies Structurally Active Compounds. Dis. Model. Mech. 2012, 5, 231–240. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Savic-Radojevic, A.; Pljesa-Ercegovac, M.; Matic, M.; Simic, D.; Radovanovic, S.; Simic, T. Novel Biomarkers of Heart Failure. Adv. Clin. Chem. 2017, 79, 93–152. [Google Scholar] [CrossRef]
- Taguchi, K.; Hirano, I.; Itoh, T.; Tanaka, M.; Miyajima, A.; Suzuki, A.; Motohashi, H.; Yamamoto, M. Nrf2 Enhances Cholangiocyte Expansion in Pten-Deficient Livers. Mol. Cell Biol. 2014, 34, 900–913. [Google Scholar] [CrossRef] [Green Version]
- Navarro-Tableros, V.; Herrera Sanchez, M.B.; Figliolini, F.; Romagnoli, R.; Tetta, C.; Camussi, G. Recellularization of Rat Liver Scaffolds by Human Liver Stem Cells. Tissue Eng. Part A 2015, 21, 1929–1939. [Google Scholar] [CrossRef] [Green Version]
- Herrera, M.B.; Fonsato, V.; Bruno, S.; Grange, C.; Gilbo, N.; Romagnoli, R.; Tetta, C.; Camussi, G. Human Liver Stem Cells Improve Liver Injury in a Model of Fulminant Liver Failure. Hepatology 2013, 57, 311–319. [Google Scholar] [CrossRef]
- Herrera, M.B.; Bruno, S.; Buttiglieri, S.; Tetta, C.; Gatti, S.; Deregibus, M.C.; Bussolati, B.; Camussi, G. Isolation and Characterization of a Stem Cell Population from Adult Human Liver. Stem Cells 2006, 24, 2840–2850. [Google Scholar] [CrossRef]
- Najimi, M.; Khuu, D.N.; Lysy, P.A.; Jazouli, N.; Abarca, J.; Sempoux, C.; Sokal, E.M. Adult-Derived Human Liver Mesenchymal-Like Cells as a Potential Progenitor Reservoir of Hepatocytes? Cell Trans. 2007, 16, 717–728. [Google Scholar] [CrossRef]







| Experiment Duration | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 0 h | 72 h | 3–6 Day | 7 Day | 8–10 Day | 11 Day | 12–14 Day | 15 Day | 16–17 Day | 18 Day | |
| Cell seeding and culture in DMEM + ROCK; coating matrigel | ||||||||||
| Medium change and dividing cells into 2 test groups and 1 control group | Medium change | |||||||||
| Control group: DMEM/F12 + KOSR + NEAA + glutamine + pen/strep + 20 ng/mL EGF, coating: matrigel | ||||||||||
| HGF group: DMEM/F12 + KOSR + NEAA + glutamine + pen/strep + DMSO + 100 ng/mL HGF, coating: matrigel | HGF group: DMEM/F12 + KOSR + NEAA + glutamine + pen/strep + dex, coating: Matrigel | |||||||||
| EGF group: DMEM/F12 + KOSR + NEAA + glutamine + pen/strep + DMSO + 100 ng/mL EGF, coating: matrigel | EGF group: DMEM/F12 + KOSR + NEAA + glutamine + pen/strep + dex, coating: Matrigel | |||||||||
| Naive hAEC analysis | Cell analysis | Cell analysis | Cell analysis | |||||||
| Gene | Full Gene Name | Group | Id Number (Thermofisher) |
|---|---|---|---|
| GAPDH | glyceraldehyde 3-phosphate dehydrogenase | Reference genes | Hs02786624_g1 |
| POLR2A | RNA polymerase II subunit A | Hs00172187_m1 | |
| PPIA | peptidylptolyl isomerase H | Hs04194521_s1 | |
| POU5F1 (OCT-4) | octamer-binding transcriptor factor 4/POU class 5 | Pluripotency related transcription factors | Hs04260367_gH |
| SOX2 | sex determining region Y-box 2 | Hs01053049_s1 | |
| PTPRC (CD45) | protein tyrosine phosphatase receptor type C | Hematopoietic stem cells marker | Hs04189704_m1 |
| DES | desmin | Mesenchymal cells marker | Hs00157258_m1 |
| EN2 | engrailed homeobox 2 | Ectodermal cells marker | Hs00171321_m1 |
| HNF4A | hepatocyte nuclear factor 4 alpha | Endodermal cells markers | Hs00230853_m1 |
| SOX17 | SRY-Box transcription factor 17 | Hs00751752_s1 | |
| AFP | alpha-fetoprotein | Hepatoblasts markers | Hs01040598_m1 |
| GSTA1 | glutathione S-transferase alpha 1 | Hs07292901_gH | |
| ALB | albumin | Hepatocytes markers | Hs00609411_m1 |
| CYP7A1 | cytochrome P450 family 7 subfamily A member 1 | Hs00167982_m1 | |
| CYP3A4 | cytochrome P450 family 3 subfamily A member 4 | Hs00604506_m1 | |
| GSTP1 | glutathione S-transferase Pi 1 | Cholangiocyte marker | Hs00943350_g1 |
| Control | EGF | HGF | |||||||
|---|---|---|---|---|---|---|---|---|---|
| 7th Day | 15th Day | 18th Day | 7th Day | 15th Day | 18th Day | 7th Day | 15th Day | 18th Day | |
| SSEA-4 | ∨ 13.27% | ∨ 87.8% | ∨ 88.9% | ∨ 4.43% | ∧ 1017.4% | ∧ 688.38% | ∧ 3.13% | ∧ 782.87% | ∧ 1017.58% |
| CD44 | ∧ 3460.29% | ∧ 3112.74% | ∧ 3623.52% | ∨ 26.66% | ∧ 12.74% | ∨ 34.12% | ∨ 29% | ∧ 0.2% | ∨ 36.08% |
| CD73 | ∧ 17.65% | ∧ 16.18% | ∧ 17.29% | ∨ 22.70% | ∨ 1.06% | ∨ 13.64% | ∨ 30.64% | ∨ 11.32% | ∨ 15.53% |
| CD90 | ∧ 2547.5% | ∧ 3205.71% | ∧ 3311.42% | ∨ 18.22% | ∨ 52.42% | ∨ 55.77% | ∨ 33.32% | ∨ 59.75% | ∨ 47.15% |
| CD105 | ∧ 973.2% | ∧ 435.48% | ∧ 504.8% | ∨ 30.46% | ∧ 114.2% | ∨ 25.44% | ∧ 13.61% | ∧ 88.15% | ∧ 63.73% |
| Phenotype | Control | EGF | HGF | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 72 h | 7th Day | 15th Day | 18th Day | 7th Day | 15th Day | 18th Day | 7th Day | 15th Day | 18th Day | ||
| SSEA-4+ | CD44+ | 2.18% | 19.75% | 0.01% f | 1.12% f | 45.36% | 71.39% a | 31.18% | 40.11% | 50.62% | 27.7% |
| CD44− | 10.52% | 4.16% i | 0.01% i | 0.02% i | 6.4% i | 11.12% i | 14.84% | 13.75% i | 15% | 39.93% a | |
| CD73+ | 0.01% | 58.68% | 47.56% | 48.51% | 43.73% | 64.38% a | 58.24% a | 39.7% | 48.76% a | 47.45% a | |
| CD73− | 0.14% | 2.15% a | 0.03% | 0.06% | 0.24% b | 2.1% ac | 2.14% ace | 4.31% abe | 3.59% ace | 4.44% abce | |
| CD90+ | 0.1% | 44.65% | 45.86% | 52.63% | 41.93% | 31.33% a | 25.93% a | 38.83% | 20.42% | 38.74% a | |
| CD90− | 0.44% | 10.71% a | 1.22% b | 1.11% b | 3.84% | 41.37% abcd | 39.13% abcd | 8.51% a | 30.17% abcd | 17.77% acd | |
| CD105+ | 2.3% | 14.66% | 0.02% a | 0.01% a | 32.69% | 67.49% | 21.88% | 33.17% | 54.47% | 61.14% | |
| CD105− | 2.73% g | 12.23% ag | 0.2% g | 1.39% g | 2.67% g | 1.36% g | 1.22% | 13.58% g | 1.98% g | 2.55% g | |
| SSEA-4− | CD44+ | 1.08% | 47.46% a | 59.93% | 78.03% a | 10.29% dj | 2.23% bd | 24.38% d | 26.46% dj | 5.55% d | 1.9% bdj |
| CD44− | 67.51% | 23.98% | 39.81% | 18.45% | 38.41% | 13.11% | 32.53% | 34.89% | 24.52% | 28% | |
| CD73+ | 46.36% | 17.91% | 43.42% | 53% | 21.82% | 20.72% | 21.42% | 14.95% a | 22.53% | 24.66% | |
| CD73− | 53.26% | 16.4% | 7.6% a | 5.7% a | 31.1% | 12.97% | 16.75% | 31.87% | 18.24% | 23.13% | |
| CD90+ | 73.81% | 10.89% a | 45.93% | 47.44% | 22.23% | 4.61% a | 7.02% a | 15.83% a | 10.23% a | 11.3% a | |
| CD90− | 20.91% | 24.18% | 3.85% | 4.18% | 28.26% | 20.94% | 25.86% | 32.04% | 38.05% | 31.58% | |
| CD105+ | 0.2% | 1.78% | 0.2%ah | 0.01% h | 0.8% | 6.26% h | 15.28% h | 5.71% | 5.94% | 5.93% | |
| CD105− | 94.11% | 68.5% | 99.82% | 98.75% | 56.03% | 23.15% acd | 60.43% | 43.8% | 34.87% cd | 28.96% acd | |
| Control | EGF | HGF | |||||||
|---|---|---|---|---|---|---|---|---|---|
| 7th Day | 15th Day | 18th Day | 7th Day | 15th Day | 18th Day | 7th Day | 15th Day | 18th Day | |
| OCT-4 | ∨ 77.53% | ∨ 74.45% | ∨ 71.8% | ∧ 9.8% | ∨ 15.51% | ∧ 56.25% | ∧ 256.86% | ∧ 29.31% | ∧ 39.06% |
| SOX2 | ∨ 99.02% | ∨ 94.75% | ∨ 94.38% | ∧ 592.3% | ∨ 37.86% | ∨ 0.04% | ∧ 5438.46% | ∧ 142.86% | ∨ 27.47% |
| DES | ∧ 20.38% | ∧ 116.67% | ∧ 858.33% | ∨ 20.69% | ∨ 53.84% | ∨ 96.52% | ∨ 17.24% | ∨ 32.69% | ∨ 94.78% |
| EN2 | ∨ 81.76% | ∨ 54.11% | ∨ 89.41% | ∧ 41.93% | ∧ 361.53% | ∧ 5.55% | ∧ 0.92% | ∧ 223.07% | ∧ 216.66% |
| SOX17 | ∨ 99.99% | ∨ 94.5% | ∨ 91.5% | ∧ 2200% | ∨ 58.62% | ∨ 86.11% | ∧ 21400% | ∧ 253.44% | ∧ 33.33% |
| CD45 | ∨ 7.33% | ∧ 420% | ∧ 33.33% | ∨ 36.43% | ∨ 94.29% | ∨ 81.5% | ∨ 28.57% | ∧ 202.91% | ∧ 135% |
| Control | EGF | HGF | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Time Point Protein/Gene | 72 h | 7th Day | 15th Day | 18th Day | 7th Day | 15th Day | 18th Day | 7th Day | 15th Day | 18th Day |
| ALB | U | ∨ 89.69% | ∨ 48.45% | ∨ 67.01% | ∨ 18% | ∨ 70% | ∧ 28.08% | ∧ 3.96% | ∧ 136% | ∧ 122.22% |
| Albumin | - | - | - | - | - | + | ++ | - | + | + |
| AFP | U | ∨ 72.5% | ∨ 90.5% | ∨ 90% | ∧ 63.63% | ∧ 21.05% | ∨ 85% | ∧ 54.54% | ∧ 400% | ∨ 2.5% |
| Alpha-fetoprotein | - | - | - | - | +++ | ++ | ++ | - | + | + |
| CYP3A4 | - | 0.00% | ∧ 1400% | ∧ 770% | 0.00% | ∨ 86% | ∨ 54.02% | 0.00% | ∧ 409.33% | ∧ 141.37% |
| CYP3A4 | X | - | - | - | - | - | + | - | + | + |
| CYP7A1 | U | ∨ 19.04% | ∧ 245.23% | ∧ 221.43% | ∧ 334.78% | ∨ 10.34% | ∨ 25.92% | ∧ 73.91% | ∧ 151.72% | ∧ 111.11% |
| CYP7A1 | - | - | - | - | ++ | + | + | + | + | ++ |
| GSTA1 | U | ∨ 99.98% | ∨ 94.33% | ∨ 93.66% | ∧ 3100% | ∨ 99.97% | ∨ 73.68% | ∧ 29900% | ∧ 100% | ∨ 62.63% |
| GSTA1 | ++++ | ++ | + | + | ++ | + | + | ++ | + | + |
| GSTP1 | U | ∧ 221.53% | ∧ 10.25% | ∧ 47.69% | ∨ 3.35% | ∧ 72.56% | ∨ 77.43% | ∨ 42.74% | ∧ 156.74% | ∧ 76.39% |
| GSTP1 | - | + | + | + | +++ | ++ | + | + | + | + |
| Cytokeratin 7 | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Michalik, M.; Wieczorek, P.; Czekaj, P. In Vitro Differentiation of Human Amniotic Epithelial Cells into Hepatocyte-like Cells. Cells 2022, 11, 2138. https://doi.org/10.3390/cells11142138
Michalik M, Wieczorek P, Czekaj P. In Vitro Differentiation of Human Amniotic Epithelial Cells into Hepatocyte-like Cells. Cells. 2022; 11(14):2138. https://doi.org/10.3390/cells11142138
Chicago/Turabian StyleMichalik, Marcin, Patrycja Wieczorek, and Piotr Czekaj. 2022. "In Vitro Differentiation of Human Amniotic Epithelial Cells into Hepatocyte-like Cells" Cells 11, no. 14: 2138. https://doi.org/10.3390/cells11142138
APA StyleMichalik, M., Wieczorek, P., & Czekaj, P. (2022). In Vitro Differentiation of Human Amniotic Epithelial Cells into Hepatocyte-like Cells. Cells, 11(14), 2138. https://doi.org/10.3390/cells11142138