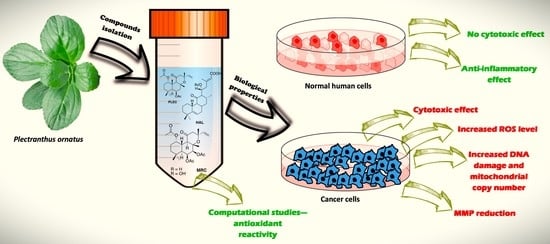
An Evaluation of the Novel Biological Properties of Diterpenes Isolated from Plectranthus ornatus Codd. In Vitro and In Silico
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Material and Isolation of Compounds from Plectranthus Ornatus
2.2. Cell Cultures
2.3. Cell Viability
2.4. Measurement of Intracellular ROS Level
2.5. Mitochondrial Membrane Potential (MMP; Δψ)
2.6. Quantitative Assessment of Mitochondrial DNA Copies
2.7. Comet Assay
2.8. mRNA Gene Expression
2.9. ADMET (Absorption, Distribution, Metabolism, Excretion, and Toxicity) Prediction
2.10. Computational Studies of Free Radical-Scavenging Properties
2.11. Molecular Docking Studies
2.12. Statistical Analyses
3. Results
3.1. Cell Viability after Treatment of Compounds HAL, PLEC and MRC
3.2. Measurement of Intracellular ROS Production after Treatment with HAL, PLEC and MRC
3.3. Mitochondrial Membrane Potential
3.4. Mitochondrial Copy Number
3.5. DNA Damage by Comet Assay
3.6. Gene Expression
3.7. ADMET Prediction
3.8. Computational Studies of Free Radical-Scavenging Properties
3.9. Molecular Docking
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sofowora, A.; Ogunbodede, E.; Onayade, A. The Role and Place of Medicinal Plants in the Strategies for Disease Prevention. Afr. J. Tradit. Complement. Altern. Med. 2013, 10, 210. [Google Scholar] [CrossRef] [PubMed]
- Yuan, H.; Ma, Q.; Ye, L.; Piao, G. The Traditional Medicine and Modern Medicine from Natural Products. Molecules 2016, 21, 559. [Google Scholar] [CrossRef] [Green Version]
- Krause, J.; Tobin, G. Discovery, Development, and Regulation of Natural Products. In Using Old Solutions to New Problems—Natural Drug Discovery in the 21st Century; IntechOpen: London, UK, 2013. [Google Scholar] [CrossRef] [Green Version]
- Barnum, C.R.; Endelman, B.J.; Shih, P.M. Utilizing Plant Synthetic Biology to Improve Human Health and Wellness. Front. Plant Sci. 2021, 12, 1824. [Google Scholar] [CrossRef]
- Kowalczyk, T.; Merecz-Sadowska, A.; Rijo, P.; Mori, M.; Hatziantoniou, S.; Górski, K.; Szemraj, J.; Piekarski, J.; Śliwiński, T.; Bijak, M.; et al. Hidden in Plants—A Review of the Anticancer Potential of the Solanaceae Family in In Vitro and In Vivo Studies. Cancers 2022, 14, 1455. [Google Scholar] [CrossRef] [PubMed]
- Merecz-Sadowska, A.; Sitarek, P.; Śliwiński, T.; Zajdel, K.; Malinowska, K.; Zielińska-Bliźniewska, H.; Kucharska, E.; Zajdel, R. In Vitro and In Silico Studies on Leonotis Nepetifolia (L.) R. Br. Root Extract against Cancer Cells. Curr. Pharm. Biotechnol. 2022, 23, 1383–1395. [Google Scholar] [CrossRef]
- Kowalczyk, T.; Merecz-Sadowska, A.; Rijo, P.; Isca, V.M.S.; Picot, L.; Wielanek, M.; Śliwiński, T.; Sitarek, P. Preliminary Phytochemical Analysis and Evaluation of the Biological Activity of Leonotis Nepetifolia (L.) R. Br Transformed Roots Extracts Obtained through Rhizobium Rhizogenes-Mediated Transformation. Cells 2021, 10, 1242. [Google Scholar] [CrossRef] [PubMed]
- Kowalczyk, T.; Sitarek, P.; Merecz-Sadowska, A.; Szyposzyńska, M.; Spławska, A.; Gorniak, L.; Bijak, M.; Śliwiński, T. Methyl Jasmonate Effect on Betulinic Acid Content and Biological Properties of Extract from Senna Obtusifolia Transgenic Hairy Roots. Molecules 2021, 26, 6208. [Google Scholar] [CrossRef] [PubMed]
- Śliwiński, T.; Sitarek, P.; Skała, E.; Isca, V.M.S.; Synowiec, E.; Kowalczyk, T.; Bijak, M.; Rijo, P. Diterpenoids from Plectranthus Spp. as Potential Chemotherapeutic Agents via Apoptosis. Pharmaceuticals 2020, 13, 123. [Google Scholar] [CrossRef]
- Sitarek, P.; Toma, M.; Ntungwe, E.; Kowalczyk, T.; Skała, E.; Wieczfinska, J.; Śliwiński, T.; Rijo, P. Insight the Biological Activities of Selected Abietane Diterpenes Isolated from Plectranthus spp. Biomolecules 2020, 10, 194. [Google Scholar] [CrossRef]
- Rice, L.J.; Brits, G.J.; Potgieter, C.J.; van Staden, J. Plectranthus: A Plant for the Future? South Afr. J. Bot. 2011, 77, 947–959. [Google Scholar] [CrossRef] [Green Version]
- Lukhoba, C.W.; Simmonds, M.S.J.; Paton, A.J. Plectranthus: A Review of Ethnobotanical Uses. J. Ethnopharmacol. 2006, 103, 1–24. [Google Scholar] [CrossRef]
- Abdel-Mogib, M.; Albar, H.A.; Batterjee, S.M. Chemistry of the Genus Plectranthus. Molecules 2002, 7, 271–301. [Google Scholar] [CrossRef] [Green Version]
- Waldia, S.; Joshi, B.C.; Pathak, U.; Joshi, M.C. The Genus Plectranthus in India and Its Chemistry. Chem. Biodivers. 2011, 8, 244–252. [Google Scholar] [CrossRef] [PubMed]
- Nascimento, F.R.; Albuquerque, K.R.S.; Oliveira, M.R.; Pizziolo, V.R.; Brasileiro, B.G.; Diaz, G.; Diaz, M.A.N. Antibiotic Activity of Plectranthus Ornatus Codd., a Traditional Medicinal Plant. An. Acad. Bras. Ciências 2017, 89, 2461–2469. [Google Scholar] [CrossRef] [Green Version]
- Ascensão, L.; Mota, L.; Castro, M.D.M. Glandular Trichomes on the Leaves and Flowers of Plectranthus Ornatus: Morphology, Distribution and Histochemistry. Ann. Bot. 1999, 84, 437–447. [Google Scholar] [CrossRef] [Green Version]
- Passinho-Soares, H.C.; Meira, P.R.; David, J.P.; Mesquita, P.R.R.; do Vale, A.E.; de Rodrigues, F.M.; de Pereira, P.A.P.; de Santana, J.R.F.; de Oliveira, F.S.; de Andrade, J.B.; et al. Volatile Organic Compounds Obtained by in Vitro Callus Cultivation of Plectranthus Ornatus Codd. (Lamiaceae). Molecules 2013, 18, 10320–10333. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rijo, P.; Gaspar-Marques, C.; Simões, M.F.; Duarte, A.; Apreda-Rojas, M.d.C.; Cano, F.H.; Rodríguez, B. Neoclerodane and Labdane Diterpenoids from Plectranthus Ornatus. J. Nat. Prod. 2002, 65, 1387–1390. [Google Scholar] [CrossRef] [PubMed]
- Rijo, P.; Gaspar-Marques, C.; Simões, M.F.; Jimeno, M.L.; Rodríguez, B. Further Diterpenoids from Plectranthus Ornatus and P. Grandidentatus. Biochem. Syst. Ecol. 2007, 35, 215–221. [Google Scholar] [CrossRef]
- Rijo, P.; Simões, M.F.; Rodríguez, B. Structural and Spectral Assignment of Three Forskolin-like Diterpenoids Isolated from Plectranthus Ornatus. Magn. Reson. Chem. 2005, 43, 595–598. [Google Scholar] [CrossRef] [PubMed]
- Sitarek, P.; Synowiec, E.; Kowalczyk, T.; Bangay, G.; Śliwiński, T.; Picot, L.; Princiotto, S.; Rijo, P. Anticancer Properties of Plectranthus ornatus-Derived Phytochemicals Inducing Apoptosis via Mitochondrial Pathway. Int. J. Mol. Sci. 2022, 23, 11653. [Google Scholar] [CrossRef]
- Reiniers, M.J.; de Haan, L.R.; Reeskamp, L.F.; Broekgaarden, M.; van Golen, R.F.; Heger, M. Analysis and Optimization of Conditions for the Use of 2′,7′-Dichlorofluorescein Diacetate in Cultured Hepatocytes. Antioxidants 2021, 10, 674. [Google Scholar] [CrossRef] [PubMed]
- Sivandzade, F.; Bhalerao, A.; Cucullo, L. Analysis of the Mitochondrial Membrane Potential Using the Cationic JC-1 Dye as a Sensitive Fluorescent Probe. Bio-Protocol 2019, 9, e3128. [Google Scholar] [CrossRef] [PubMed]
- Bijak, M.; Synowiec, E.; Sitarek, P.; Sliwiński, T.; Saluk-Bijak, J. Evaluation of the Cytotoxicity and Genotoxicity of Flavonolignans in Different Cellular Models. Nutrients 2017, 9, 1356. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ceremuga, M.; Stela, M.; Janik, E.; Gorniak, L.; Synowiec, E.; Sliwinski, T.; Sitarek, P.; Saluk-Bijak, J.; Bijak, M. Melittin—A Natural Peptide from Bee Venom Which Induces Apoptosis in Human Leukaemia Cells. Biomolecules 2020, 10, 247. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sitarek, P.; Synowiec, E.; Kowalczyk, T.; Śliwiński, T.; Skała, E. An In Vitro Estimation of the Cytotoxicity and Genotoxicity of Root Extract from Leonurus Sibiricus L. Overexpressing AtPAP1 against Different Cancer Cell Lines. Molecules 2018, 23, 2049. [Google Scholar] [CrossRef] [Green Version]
- Singh, N.P.; McCoy, M.T.; Tice, R.R.; Schneider, E.L. A Simple Technique for Quantitation of Low Levels of DNA Damage in Individual Cells. Exp. Cell Res. 1988, 175, 184–191. [Google Scholar] [CrossRef] [Green Version]
- Klaude, M.; Eriksson, S.; Nygren, J.; Ahnström, G. The Comet Assay: Mechanisms and Technical Considerations. Mutat. Res. 1996, 363, 89–96. [Google Scholar] [CrossRef]
- Smith, C.C.; O’Donovan, M.R.; Martin, E.A. HOGG1 Recognizes Oxidative Damage Using the Comet Assay with Greater Specificity than FPG or ENDOIII. Mutagenesis 2006, 21, 185–190. [Google Scholar] [CrossRef] [Green Version]
- Schmittgen, T.D.; Livak, K.J. Analyzing Real-Time PCR Data by the Comparative CT Method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef] [PubMed]
- Snyder, H.D.; Kucukkal, T.G. Computational Chemistry Activities with Avogadro and ORCA. J. Chem. Educ. 2021, 98, 1335–1341. [Google Scholar] [CrossRef]
- Hunter, A.D. ACD/ChemSketch 1.0 (Freeware); ACD/ChemSketch 2.0 and Its Tautomers, Dictionary, and 3D Plug-Ins; ACD/HNMR 2.0; ACD/CNMR 2.0. J. Chem. Educ. 1997, 74, 905. [Google Scholar] [CrossRef] [Green Version]
- O’Boyle, N.M.; Banck, M.; James, C.A.; Morley, C.; Vandermeersch, T.; Hutchison, G.R. Open Babel: An Open Chemical Toolbox. J. Cheminform. 2011, 3, 33. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morris, G.M.; Ruth, H.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AutoDock4 and AutoDockTools4: Automated Docking with Selective Receptor Flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morris, G.M.; Goodsell, D.S.; Halliday, R.S.; Huey, R.; Hart, W.E.; Belew, R.K.; Olson, A.J. Automated Docking Using a Lamarckian Genetic Algorithm and an Empirical Binding Free Energy Function. J. Comput. Chem. 1998, 19, 1639–1662. [Google Scholar] [CrossRef]
- Coimbra, J.T.S.; Feghali, R.; Ribeiro, R.P.; Ramos, M.J.; Fernandes, P.A. The Importance of Intramolecular Hydrogen Bonds on the Translocation of the Small Drug Piracetam through a Lipid Bilayer. RSC Adv. 2021, 11, 899–908. [Google Scholar] [CrossRef]
- Daina, A.; Michielin, O.; Zoete, V. SwissADME: A Free Web Tool to Evaluate Pharmacokinetics, Drug-likeness and Medicinal Chemistry Friendliness of Small Molecules. Sci. Rep. 2017, 7, 42717. [Google Scholar] [CrossRef] [Green Version]
- Seelig, A. P-Glycoprotein: One Mechanism, Many Tasks and the Consequences for Pharmacotherapy of Cancers. Front. Oncol. 2020, 10, 576559. [Google Scholar] [CrossRef]
- Purnapatre, K.; Khattar, S.K.; Saini, K.S. Cytochrome P450s in the development of target-based anticancer drugs. Cancer Lett. 2008, 259, 1–15. [Google Scholar] [CrossRef]
- Veeresham, C. Natural Products Derived from Plants as a Source of Drugs. J. Adv. Pharm. Technol. Res. 2012, 3, 200–201. [Google Scholar] [CrossRef]
- Lautié, E.; Russo, O.; Ducrot, P.; Boutin, J.A. Unraveling Plant Natural Chemical Diversity for Drug Discovery Purposes. Front. Pharmacol. 2020, 11, 397. [Google Scholar] [CrossRef]
- Atanasov, A.G.; Waltenberger, B.; Pferschy-Wenzig, E.M.; Linder, T.; Wawrosch, C.; Uhrin, P.; Temml, V.; Wang, L.; Schwaiger, S.; Heiss, E.H.; et al. Discovery and Resupply of Pharmacologically Active Plant-Derived Natural Products: A Review. Biotechnol. Adv. 2015, 33, 1582–1614. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Howes, M.J.R.; Quave, C.L.; Collemare, J.; Tatsis, E.C.; Twilley, D.; Lulekal, E.; Farlow, A.; Li, L.; Cazar, M.E.; Leaman, D.J.; et al. Molecules from Nature: Reconciling Biodiversity Conservation and Global Healthcare Imperatives for Sustainable Use of Medicinal Plants and Fungi. Plants People Planet 2020, 2, 463–481. [Google Scholar] [CrossRef]
- Pan, S.Y.; Zhou, S.F.; Gao, S.H.; Yu, Z.L.; Zhang, S.F.; Tang, M.K.; Sun, J.N.; Ma, D.L.; Han, Y.F.; Fong, W.F.; et al. New Perspectives on How to Discover Drugs from Herbal Medicines: CAM’s Outstanding Contribution to Modern Therapeutics. Evid. Based Complement. Altern. Med. 2013, 2013, 627375. [Google Scholar] [CrossRef] [Green Version]
- Bhat, S.G. Medicinal Plants and Its Pharmacological Values. In Natural Medicinal Plants; IntechOpen: London, UK, 2021. [Google Scholar] [CrossRef]
- Stéphane, F.F.Y.; Jules, B.K.J.; Batiha, G.E.-S.; Ali, I.; Bruno, L.N. Extraction of Bioactive Compounds from Medicinal Plants and Herbs. In Natural Medicinal Plants; IntechOpen: London, UK, 2021. [Google Scholar] [CrossRef]
- Demetzos, C.; Dimas, K.S. Labdane-Type Diterpenes: Chemistry and Biological Activity. Stud. Nat. Prod. Chem. 2001, 25, 235–292. [Google Scholar] [CrossRef]
- Mani, V.; Park, S.; Kim, J.A.; Lee, S.I.; Lee, K. Metabolic Perturbation and Synthetic Biology Strategies for Plant Terpenoid Production—An Updated Overview. Plants 2021, 10, 2179. [Google Scholar] [CrossRef]
- Silva, L.; Gomes, A.C. Diterpene Lactones with Labdane, Halimane and Clerodane Frameworks. Nat. Prod. Commun. 2011, 6, 497–504. [Google Scholar] [CrossRef] [Green Version]
- Roncero, A.M.; Tobal, I.E.; Moro, R.F.; Díez, D.; Marcos, I.S. Halimane Diterpenoids: Sources, Structures, Nomenclature and Biological Activities. Nat. Prod. Rep. 2018, 35, 955–991. [Google Scholar] [CrossRef]
- Majhi, S. Diterpenoids: Natural Distribution, Semisynthesis at Room Temperature and Pharmacological Aspects—A Decade Update. ChemistrySelect 2020, 5, 12450–12464. [Google Scholar] [CrossRef]
- Saha, P.; Rahman, F.I.; Hussain, F.; Rahman, S.M.A.; Rahman, M.M. Antimicrobial Diterpenes: Recent Development From Natural Sources. Front. Pharmacol. 2022, 12, 820312. [Google Scholar] [CrossRef]
- Seaman, F.; Bohlmann, F.; Zdero, C.; Mabry, T.J. Biological Activity of Diterpenes. In Diterpenes Flower. Plants; Springer: Berlin/Heidelberg, Germany, 1990; pp. 485–492. [Google Scholar] [CrossRef]
- Sánchez, M.; Mazzuca, M.; Veloso, M.J.; Fernández, L.R.; Siless, G.; Puricelli, L.; Palermo, J.A. Cytotoxic Terpenoids from Nardophyllum Bryoides. Phytochemistry 2010, 71, 1395–1399. [Google Scholar] [CrossRef]
- Silva, C.G.; Santos, H.M.; Barbosa, J.P.; Costa, G.L.; Rodrigues, F.A.R.; Oliveira, D.F.; Costa-Lotufo, L.V.; Alves, R.J.V.; Eleutherio, E.C.A.; Rezende, C.M. Structure Elucidation, Antimicrobial and Cytotoxic Activities of a Halimane Isolated from Vellozia Kolbekii Alves (Velloziaceae). Chem. Biodivers. 2015, 12, 1891–1901. [Google Scholar] [CrossRef] [PubMed]
- Abdel-Kader, M.; Berger, J.M.; Slebodnick, C.; Hoch, J.; Malone, S.; Wisse, J.H.; Werkhoven, M.C.M.; Mamber, S.; Kingston, D.G.I. Isolation and Absolute Configuration of Ent-Halimane Diterpenoids from Hymenaea Courbaril from the Suriname Rain Forest1. J. Nat. Prod. 2001, 65, 11–15. [Google Scholar] [CrossRef]
- Zorova, L.D.; Popkov, V.A.; Plotnikov, E.Y.; Silachev, D.N.; Pevzner, I.B.; Jankauskas, S.S.; Babenko, V.A.; Zorov, S.D.; Balakireva, A.V.; Juhaszova, M.; et al. Mitochondrial Membrane Potential. Anal. Biochem. 2018, 552, 50–59. [Google Scholar] [CrossRef] [PubMed]
- Kühlbrandt, W. Structure and Function of Mitochondrial Membrane Protein Complexes. BMC Biol. 2015, 13, 89. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, C.; Youle, R.J. The Role of Mitochondria in Apoptosis*. Annu. Rev. Genet. 2009, 43, 95–118. [Google Scholar] [CrossRef] [Green Version]
- Burke, P.J. Mitochondria, Bioenergetics and Apoptosis in Cancer. Trends Cancer 2017, 3, 857–870. [Google Scholar] [CrossRef]
- Hu, L.; Yao, X.; Shen, Y. Altered Mitochondrial DNA Copy Number Contributes to Human Cancer Risk: Evidence from an Updated Meta-Analysis. Sci. Rep. 2016, 6, 35895. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roos, W.P.; Kaina, B. DNA Damage-Induced Cell Death by Apoptosis. Trends Mol. Med. 2006, 12, 440–450. [Google Scholar] [CrossRef] [PubMed]
- Spiegel, M. Current Trends in Computational Quantum Chemistry Studies on Antioxidant Radical Scavenging Activity. J. Chem. Inf. Model. 2022, 2022, 2639–2658. [Google Scholar] [CrossRef] [PubMed]
- Ji, L.; Liu, T.; Liu, J.; Chen, Y.; Wang, Z. Andrographolide Inhibits Human Hepatoma-Derived Hep3B Cell Growth through the Activation of c-Jun N-Terminal Kinase. Planta Med. 2007, 73, 1397–1401. [Google Scholar] [CrossRef] [PubMed]
- Luo, P.; Yu, Q.; Liu, S.N.; Xia, W.J.; Fang, Y.Y.; An, L.K.; Gu, Q.; Xu, J. Diterpenoids with Diverse Scaffolds from Vitex Trifolia as Potential Topoisomerase I Inhibitor. Fitoterapia 2017, 120, 108–116. [Google Scholar] [CrossRef]
- Chen, L.; Deng, H.; Cui, H.; Fang, J.; Zuo, Z.; Deng, J.; Li, Y.; Wang, X.; Zhao, L. Inflammatory Responses and Inflammation-Associated Diseases in Organs. Oncotarget 2017, 9, 7204–7218. [Google Scholar] [CrossRef] [Green Version]
- Das, U.N. Inflammation. In Molecular Basis of Health and Disease; Springer: Berlin/Heidelberg, Germany, 2011; pp. 15–100. [Google Scholar] [CrossRef]
- Kany, S.; Vollrath, J.T.; Relja, B. Cytokines in Inflammatory Disease. Int. J. Mol. Sci. 2019, 20, 6008. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parameswaran, N.; Patial, S. Tumor Necrosis Factor-α Signaling in Macrophages. Crit. Rev. Eukaryot. Gene Expr. 2010, 20, 87–103. [Google Scholar] [CrossRef]
- McQualter, J.L.; Darwiche, R.; Ewing, C.; Onuki, M.; Kay, T.W.; Hamilton, J.A.; Reid, H.H.; Bernard, C.C.A. Granulocyte Macrophage Colony-Stimulating Factor: A New Putative Therapeutic Target in Multiple Sclerosis. J. Exp. Med. 2001, 194, 873–881. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Hunt, N.H.; Bao, S. The Role of Granulocyte Macrophage-Colony-Stimulating Factor in Acute Intestinal Inflammation. Cell Res. 2008, 18, 1220–1229. [Google Scholar] [CrossRef] [PubMed]
- Repositório Da Universidade de Lisboa: Phytochemical Study and Biological Activities of Diterpenes and Derivatives from Plectranthus Species. Available online: https://repositorio.ul.pt/handle/10451/2833 (accessed on 9 July 2022).
- Chiadak, J.D.; Arsenijevic, T.; Verstrepen, K.; Gregoire, F.; Bolaky, N.; Delforge, V.; Flamand, V.; Perret, J.; Delporte, C. Forskolin Inhibits Lipopolysaccharide-Induced Modulation of MCP-1 and GPR120 in 3T3-L1 Adipocytes through an Inhibition of NFκ B. Mediators Inflamm. 2016, 2016, 1431789. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karthika, K.; Jamuna, S.; Abinaya, G.; Venkatachalapathi, A.; Thenmozhi, K.; Paulsamy, S. Evaluation of Anti-Inflammatory and Antioxidant Properties of Crude Extract and Forskolin from Solena Amplexicaulis Leaf. Indian J. Pharm. Sci. 2016, 78, 377–387. [Google Scholar] [CrossRef]
- Li, H.Y.; Li, Y.; Wei, W.J.; Ma, K.L.; Chen, J.J.; Gao, K. Halimane and Labdane Diterpenoids from Leonurus Japonicus and Their Anti-Inflammatory Activity. Phytochemistry 2020, 172, 112280. [Google Scholar] [CrossRef] [PubMed]
- Malik, A.; Naz, A.; Ahmad, S.; Hafeez, M.; Awan, F.M.; Jafar, T.H.; Zahid, A.; Ikram, A.; Rauff, B.; Hassan, M. Inhibitory Potential of Phytochemicals on Interleukin-6-Mediated T-Cell Reduction in COVID-19 Patients: A Computational Approach. Bioinform. Biol. Insights 2021, 15, 11779322211021430. [Google Scholar] [CrossRef]
- Prathap, L.; Jayaraman, S.; Roy, A.; Santhakumar, P.; Jeevitha, M. Molecular docking analysis of stachydrine and sakuranetin with IL-6 and TNF-α in the context of inflammation. Bioinformation 2021, 17, 363–368. [Google Scholar] [CrossRef] [PubMed]
- Shen, L.; Jiang, Y.; Lu, J.; Wang, G.; Zhang, X.; He, S.; Wang, C.; Li, Z. Molecular Mechanism of Jinchan Oral Liquid in the Treatment of Children with Respiratory Syncytial Virus Pneumonia Based on Network Pharmacology and Molecular Docking Technology. Biomed Res. Int. 2021, 2021, 6471400. [Google Scholar] [CrossRef] [PubMed]
- Alzahrani, A.A. New Investigation into the Molecular Mechanism of Andrographolide towards Reducing Cytokine Storm. Molecules 2022, 27, 4555. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Du, X.; Liu, Y.; Liu, Q.Q.; Zhi, W.B.; Wang, C.L.; Zhou, J.; Li, Y.; Zhang, H. A Systems Pharmacology Approach for Identifying the Multiple Mechanisms of Action for the Rougui-Fuzi Herb Pair in the Treatment of Cardiocerebral Vascular Diseases. Evid. Based Complement. Altern. Med. 2020, 2020, 5196302. [Google Scholar] [CrossRef] [PubMed]










| Compound Name | HAL | PLEC | 1,6-di-O-acetylforskolin | 1,6-do-O-acetyl-9-deoxyforskolin | Gemcitabine |
|---|---|---|---|---|---|
| Formula | C25H36O5 | C24H36O6 | C26H38O8 | C26H38O9 | C9H11F2N3O4 |
| MW 1 (g/mol) | 416.55 | 420.54 | 478.58 | 494.57 | 236.20 |
| RB 2 | 6 | 5 | 7 | 7 | 2 |
| HBA 3 | 5 | 6 | 8 | 9 | 7 |
| HBD 4 | 1 | 0 | 0 | 1 | 3 |
| Fraction C sp 3,5 | 0.72 | 0.79 | 0.77 | 0.77 | 0.56 |
| TPSA 6 (Å2) | 80.67 | 78.90 | 105.20 | 125.43 | 110.60 |
| Log Po/w 7 | 4.22 | 3.60 | 3.31 | 2.68 | −1.46 |
| LogS 8 | −5.32 | −4.21 | −4.18 | −3.80 | −0.67 |
| Lipinski 9 | Yes (0) | Yes (0) | Yes (0) | Yes (0) | Yes (0) |
| Bioavailability Score 10 | 0.56 | 0.55 | 0.55 | 0.55 | 0.55 |
| Compound Name | HAL | PLEC | 1,6-di-O-acetylforskolin | 1,6-do-O-acetyl-9-deoxyforskolin | Gemcitabine |
|---|---|---|---|---|---|
| GI absorption 1 | High | High | High | High | High |
| BBB Permeant 2 | No | No | No | No | No |
| P-gp Substrate 3 | Yes | No | No | Yes | No |
| CYP1A2 inhibitor | No | No | No | No | No |
| CYP2C19 inhibitor | No | No | No | No | No |
| CYP2C9 inhibitor | Yes | No | No | No | No |
| CYP2D6 inhibitor | No | No | No | No | No |
| CYP3A4 inhibitor | Yes | No | No | Yes | No |
| Log Kp 4 (cm/s) | −5.13 | −6.51 | −7.15 | −7.79 | −8.94 |
| LD50 5 (mg/kg) | 3300 | 100 | 2550 | 2550 | 1000 |
| Hepatotoxicity | |||||
| Carcinogenicity | |||||
| Mutagenicity | |||||
| A 6 | |||||
| p 7 | 0.88 | 0.82 | 0.75 | 0.68 | 0.81 |
| Cytotoxicity | |||||
| A 6 | |||||
| p 7 | 0.80 | 0.78 | 0.69 | 0.72 | 0.94 |
| Molecular Descriptor | HAL | 1,6-di-O-acetylforskolin | 1,6-di-O-acetyl-9-deoxyforskolin | PLEC | Caffeic Acid (Positive Standard) | Phenol (Negative Standard) |
|---|---|---|---|---|---|---|
| HOMO energy (eV) | −6.399 | −6.959 | −6.782 | −6.892 | −6.186 | −6.238 |
| LUMO energy (eV) | −1.605 | −1.109 | −1.113 | −1.013 | −2.059 | −0.377 |
| Egaps (eV) | 4.794 | 5.850 | 5.669 | 5.879 | 4.127 | 5.861 |
| IP (eV) | 6.399 | 6.959 | 6.782 | 6.892 | 6.186 | 6.238 |
| EA (eV) | 1.605 | 1.109 | 1.113 | 1.103 | 2.059 | 0.377 |
| Proteins | Plant-Derived Compounds | Binding Free Energy (kcal/mol) | Hydrogen Bonding | Other Interactions |
|---|---|---|---|---|
| IL-6 | HAL | −10.33 | GLU A:42 ARG A:104 GLU A:106 ASP A:160 | LYS A:46 PHE A:105 THR A:163 |
| 1,6-di-O-acetylforskolin | −10.87 | - | TYR A:31 ILE A:32 ASP A:34 GLN A:111 VAL A:115 | |
| 1,6-di-O-acetyl-9-deoxyforskolin | −12.16 | - | LYS A:66 GLU A:172 SER A:169 | |
| PLEC | −11.06 | - | PRO A:65 SER A:169 GLU A:172 | |
| IL-8 | HAL | −14.48 | ILE A:8 LYS A:9 LEU A:47 CYS A:48 | TYR A:11 GLU A:46 |
| 1,6-di-O-acetylforskolin | −16.26 | ILE A:8 LYS A:9 | TYR A:11 GLU A:46 LEU A:47 | |
| 1,6-di-O-acetyl-9-deoxyforskolin | −15.85 | ILE A:8 LYS A:9 ARG A:45 | GLU A:46 | |
| PLEC | −14.82 | ILE A:8 LYS A:9 GLU A:46 | TYR A:11 LEU A:47 | |
| TNF-α | HAL | −11.96 | - | GLU A:116 PRO C:100 GLU C:116 |
| 1,6-di-O-acetylforskolin | −14.12 | - | GLU A:116 LYS C:98 | |
| -di-O-acetyl-9-deoxyforskolin) | −15.18 | GLN A:102 SER B:99 GLN C:102 | CYS A:101 GLU A:116 GLU B:116 LYS C:98 TYR C:115 GLU C:116 | |
| PLEC | −13.49 | CYS A:69 GLN B:102 GLN C:102 | GLU C:116 | |
| GM-CSF | HAL | −11.36 | TRP B13 GLU B:14 SER B:82 HIS B:83 GLN B:86 | PRO B:8 PRO B:6 PRO B:12 HIS B:87 |
| 1,6-di-O-acetylforskolin | −13.20 | GLN A:86 | PRO A:8 PRO A:12 GLU A:14 HIS A:87 | |
| 1,6-di-O-acetyl-9-deoxyforskolin) | −13.84 | TRP A:13 GLN A:86 | SER A:8 GLU A:14 SER A:82 HIS A:87 | |
| PLEC | −12.07 | GLN A:86 | PRO A:8 PRO A:12 GLU A:14 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sitarek, P.; Kowalczyk, T.; Synowiec, E.; Merecz-Sadowska, A.; Bangay, G.; Princiotto, S.; Śliwiński, T.; Rijo, P. An Evaluation of the Novel Biological Properties of Diterpenes Isolated from Plectranthus ornatus Codd. In Vitro and In Silico. Cells 2022, 11, 3243. https://doi.org/10.3390/cells11203243
Sitarek P, Kowalczyk T, Synowiec E, Merecz-Sadowska A, Bangay G, Princiotto S, Śliwiński T, Rijo P. An Evaluation of the Novel Biological Properties of Diterpenes Isolated from Plectranthus ornatus Codd. In Vitro and In Silico. Cells. 2022; 11(20):3243. https://doi.org/10.3390/cells11203243
Chicago/Turabian StyleSitarek, Przemysław, Tomasz Kowalczyk, Ewelina Synowiec, Anna Merecz-Sadowska, Gabrielle Bangay, Salvatore Princiotto, Tomasz Śliwiński, and Patricia Rijo. 2022. "An Evaluation of the Novel Biological Properties of Diterpenes Isolated from Plectranthus ornatus Codd. In Vitro and In Silico" Cells 11, no. 20: 3243. https://doi.org/10.3390/cells11203243
APA StyleSitarek, P., Kowalczyk, T., Synowiec, E., Merecz-Sadowska, A., Bangay, G., Princiotto, S., Śliwiński, T., & Rijo, P. (2022). An Evaluation of the Novel Biological Properties of Diterpenes Isolated from Plectranthus ornatus Codd. In Vitro and In Silico. Cells, 11(20), 3243. https://doi.org/10.3390/cells11203243