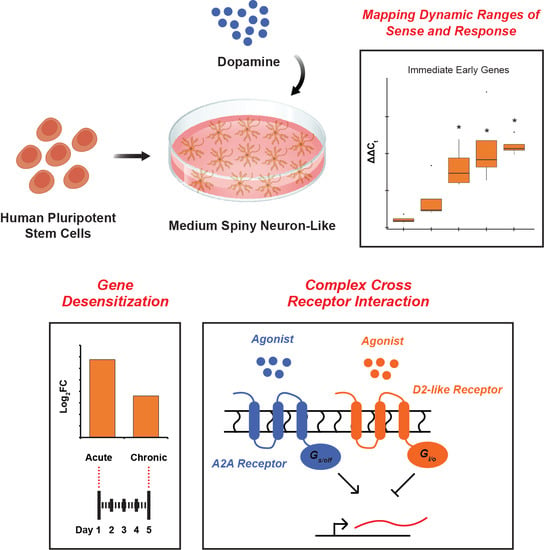
Human Pluripotent Stem Cell-Derived Medium Spiny Neuron-like Cells Exhibit Gene Desensitization
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Cultures
2.2. Dosing Experiments
2.3. Immunocytochemistry
2.4. Immunostaining Quantification
2.5. RNA Extraction and QRT-PCR
2.6. RNA-Sequencing and Analysis
2.7. Statistical Analysis
3. Results
3.1. hPSC-Derived Neurons Express Markers of MSNs
3.2. hMSN-like Cells Exhibit Dose and Time-Dependent Responses to Dopamine
3.3. Chronic Administration of Dopamine Leads to Desensitization of Genes Implicated in Cocaine and Dopamine Responses
3.4. Time Course of Chronic Dopamine Administration Reveals Peak in DEGs at Day 3 and Desensitization at Day 5
3.5. hMSN-like Cells Capture Some Features of Dopamine Receptor Cross-Interactions
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Maze, I.; Iii, H.E.C.; Dietz, D.M.; Laplant, Q.; Renthal, W.; Russo, S.J.; Mechanic, M.; Mouzon, E.; Neve, R.L.; Stephen, J.; et al. Essential Role of the Histone Methyltransferase G9a in Cocaine Induced Plasticity. Science 2010, 327, 213–216. [Google Scholar] [CrossRef] [Green Version]
- Renthal, W.; Carle, T.L.; Maze, I.; Covington, H.E.; Truong, H.-T.; Alibhai, I.; Kumar, A.; Montgomery, R.L.; Olson, E.N.; Nestler, E.J. ΔFosB Mediates Epigenetic Desensitization of the C-Fos Gene After Chronic Amphetamine Exposure. J. Neurosci. 2008, 28, 7344–7349. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rusconi, F.; Battaglioli, E. Acute Stress-Induced Epigenetic Modulations and Their Potential Protective Role Toward Depression. Front. Mol. Neurosci. 2018, 11, 184. [Google Scholar] [CrossRef] [PubMed]
- Nestler, E.J.; Barrot, M.; Self, D.W. FosB: A Sustained Molecular Switch for Addiction. Proc. Natl. Acad. Sci. USA 2001, 98, 11042–11046. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tang, T.-S.; Slow, E.; Lupu, V.; Stavrovskaya, I.G.; Sugimori, M.; Llinás, R.; Kristal, B.S.; Hayden, M.R.; Bezprozvanny, I. Disturbed Ca2+ Signaling and Apoptosis of Medium Spiny Neurons in Huntington’s Disease. Proc. Natl. Acad. Sci. USA 2005, 102, 2602–2607. [Google Scholar] [CrossRef] [Green Version]
- Deutch, A.Y.; Colbran, R.J.; Winder, D.J. Striatal Plasticity and Medium Spiny Neuron Dendritic Remodeling in Parkinsonism. Parkinsonism Relat. Disord. 2007, 13, S251–S258. [Google Scholar] [CrossRef] [Green Version]
- Chandra, R.; Lobo, M.K. Beyond Neuronal Activity Markers: Select Immediate Early Genes in Striatal Neuron Subtypes Functionally Mediate Psychostimulant Addiction. Front. Behav. Neurosci. 2017, 11, 112. [Google Scholar] [CrossRef] [Green Version]
- Le Cann, K.; Foerster, A.; Rösseler, C.; Erickson, A.; Hautvast, P.; Giesselmann, S.; Pensold, D.; Kurth, I.; Rothermel, M.; Mattis, V.B.; et al. The Difficulty to Model Huntington’s Disease in Vitro Using Striatal Medium Spiny Neurons Differentiated from Human Induced Pluripotent Stem Cells. Sci. Rep. 2021, 11, 6934. [Google Scholar] [CrossRef]
- Lissek, T.; Andrianarivelo, A.; Saint-Jour, E.; Allichon, M.-C.; Bauersachs, H.G.; Nassar, M.; Piette, C.; Pruunsild, P.; Tan, Y.-W.; Forget, B.; et al. Npas4 Regulates Medium Spiny Neuron Physiology and Gates Cocaine-Induced Hyperlocomotion. EMBO Rep. 2021, 22, e51882. [Google Scholar] [CrossRef]
- Yuen, J.; Goyal, A.; Rusheen, A.E.; Kouzani, A.Z.; Berk, M.; Kim, J.H.; Tye, S.J.; Blaha, C.D.; Bennet, K.E.; Jang, D.-P.; et al. Cocaine-Induced Changes in Tonic Dopamine Concentrations Measured Using Multiple-Cyclic Square Wave Voltammetry in Vivo. Front. Pharmacol. 2021, 12, 1710. [Google Scholar] [CrossRef]
- Heien, M.L.A.V.; Khan, A.S.; Ariansen, J.L.; Cheer, J.F.; Phillips, P.E.M.; Wassum, K.M.; Wightman, R.M. Real-Time Measurement of Dopamine Fluctuations after Cocaine in the Brain of Behaving Rats. Proc. Natl. Acad. Sci. USA 2005, 102, 10023–10028. [Google Scholar] [CrossRef] [Green Version]
- Mishra, A.; Singh, S.; Shukla, S. Physiological and Functional Basis of Dopamine Receptors and Their Role in Neurogenesis: Possible Implication for Parkinson’s Disease. J. Exp. Neurosci. 2018, 12, 1179069518779829. [Google Scholar] [CrossRef] [Green Version]
- Diana, M. The Dopamine Hypothesis of Drug Addiction and Its Potential Therapeutic Value. Front. Psychiatry 2011, 2, 64. [Google Scholar] [CrossRef] [Green Version]
- Kumar, A.; Choi, K.H.; Renthal, W.; Tsankova, N.M.; Theobald, D.E.H.; Truong, H.T.; Russo, S.J.; LaPlant, Q.; Sasaki, T.S.; Whistler, K.N.; et al. Chromatin Remodeling Is a Key Mechanism Underlying Cocaine-Induced Plasticity in Striatum. Neuron 2005, 48, 303–314. [Google Scholar] [CrossRef] [Green Version]
- Nestler, E. The Neurobiology of Cocaine Addiction. Sci. Pract. Perspect. 2005, 3, 4–10. [Google Scholar] [CrossRef] [Green Version]
- Dudman, J.T.; Gerfen, C.R. Chapter 17—The Basal Ganglia. In Paxinos, G.B.T.-T.R.N.S, 4th ed.; Academic Press: San Diego, CA, USA, 2015; pp. 391–440. ISBN 978-0-12-374245-2. [Google Scholar]
- Arber, C.; Precious, S.V.; Cambray, S.; Risner-Janiczek, J.R.; Kelly, C.; Noakes, Z.; Fjodorova, M.; Heuer, A.; Ungless, M.A.; Rodriguez, T.A.; et al. Activin A Directs Striatal Projection Neuron Differentiation of Human Pluripotent Stem Cells. Development 2015, 142, 1375–1386. [Google Scholar] [CrossRef] [Green Version]
- Golas, M.M. Human Cellular Models of Medium Spiny Neuron Development and Huntington Disease. Life Sci. 2018, 209, 179–196. [Google Scholar] [CrossRef]
- Carri, A.D.; Onorati, M.; Lelos, M.J.; Castiglioni, V.; Faedo, A.; Menon, R.; Camnasio, S.; Vuono, R.; Spaiardi, P.; Talpo, F.; et al. Developmentally Coordinated Extrinsic Signals Drive Human Pluripotent Stem Cell Differentiation toward Authentic DARPP-32+ Medium-Sized Spiny Neurons. Development 2013, 140, 301–312. [Google Scholar] [CrossRef] [Green Version]
- Hunt, C.P.J.; Pouton, C.W.; Haynes, J.M. Characterising the Developmental Profile of Human Embryonic Stem Cell-Derived Medium Spiny Neuron Progenitors and Assessing Mature Neuron Function Using a CRISPR-Generated Human DARPP-32WT/EGFP-AMPreporter Line. Neurochem. Int. 2017, 106, 3–13. [Google Scholar] [CrossRef]
- Iannielli, A.; Ugolini, G.S.; Cordiglieri, C.; Bido, S.; Rubio, A.; Colasante, G.; Valtorta, M.; Cabassi, T.; Rasponi, M.; Broccoli, V. Reconstitution of the Human Nigro-Striatal Pathway on-a-Chip Reveals OPA1-Dependent Mitochondrial Defects and Loss of Dopaminergic Synapses. Cell Rep. 2019, 29, 4646–4656.e4. [Google Scholar] [CrossRef] [Green Version]
- Grigor’eva, E.V.; Malankhanova, T.B.; Surumbayeva, A.; Pavlova, S.V.; Minina, J.M.; Kizilova, E.A.; Suldina, L.A.; Morozova, K.N.; Kiseleva, E.; Sorokoumov, E.D.; et al. Generation of GABAergic Striatal Neurons by a Novel IPSC Differentiation Protocol Enabling Scalability and Cryopreservation of Progenitor Cells. Cytotechnology 2020, 72, 649–663. [Google Scholar] [CrossRef] [PubMed]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An Open-Source Platform for Biological-Image Analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sen, D.; Voulgaropoulos, A.; Drobna, Z.; Keung, A.J. Human Cerebral Organoids Reveal Early Spatiotemporal Dynamics and Pharmacological Responses of UBE3A. Stem Cell Rep. 2020, 15, 845–854. [Google Scholar] [CrossRef] [PubMed]
- Untergasser, A.; Cutcutache, I.; Koressaar, T.; Ye, J.; Faircloth, B.C.; Remm, M.; Rozen, S.G. Primer3--New Capabilities and Interfaces. Nucleic Acids Res. 2012, 40, e115. [Google Scholar] [CrossRef] [Green Version]
- Krämer, A.; Green, J.; Pollard, J., Jr.; Tugendreich, S. Causal Analysis Approaches in Ingenuity Pathway Analysis. Bioinformatics 2014, 30, 523–530. [Google Scholar] [CrossRef]
- Raudvere, U.; Kolberg, L.; Kuzmin, I.; Arak, T.; Adler, P.; Peterson, H.; Vilo, J. G:Profiler: A Web Server for Functional Enrichment Analysis and Conversions of Gene Lists (2019 Update). Nucleic Acids Res. 2019, 47, W191–W198. [Google Scholar] [CrossRef] [Green Version]
- Allaire, J. Rstudio: Integrated Development Environment for R. 2012, Volume 770, pp. 165–171. Available online: https://www.r-project.org/conferences/useR-2011/abstracts/180111-allairejj.pdf (accessed on 25 March 2022).
- Hulsen, T. BioVenn—An R and Python Package for the Comparison and Visualization of Biological Lists Using Area-Proportional Venn Diagrams. Data Sci. 2021, 4, 51–61. [Google Scholar] [CrossRef]
- VIB-UGENT for Plant Systems Biology Calculate and Draw Custom Venn Diagrams. Available online: http://bioinformatics.psb.ugent.be/webtools/Venn/ (accessed on 12 January 2022).
- Wickham, H. Ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2016; ISBN 978-3-319-24277-4. [Google Scholar]
- Gokce, O.; Stanley, G.M.; Treutlein, B.; Neff, N.F.; Camp, J.G.; Malenka, R.C.; Rothwell, P.E.; Fuccillo, M.V.; Südhof, T.C.; Quake, S.R. Cellular Taxonomy of the Mouse Striatum as Revealed by Single-Cell RNA-Seq. Cell Rep. 2016, 16, 1126–1137. [Google Scholar] [CrossRef] [Green Version]
- Straccia, M.; Barriga, G.G.D.; Sanders, P.; Bombau, G.; Carrere, J.; Mairal, P.B.; Vinh, N.N.; Yung, S.; Kelly, C.M.; Svendsen, C.N.; et al. Quantitative High-Throughput Gene Expression Profiling of Human Striatal Development to Screen Stem Cell-Derived Medium Spiny Neurons. Mol. Ther. -Methods Clin. Dev. 2015, 2, 1–15. [Google Scholar] [CrossRef]
- Benarroch, E.E. Effects of Acetylcholine in the Striatum. Recent Insights and Therapeutic Implications. Neurology 2012, 79, 274–281. [Google Scholar] [CrossRef]
- Garris, P.A.; Ciolkowski, E.L.; Pastore, P.; Wightman, R.M. Efflux of Dopamine from the Synaptic Cleft in the Nucleus Accumbens of the Rat Brain. J. Neurosci. 1994, 14, 6084–6093. [Google Scholar] [CrossRef] [Green Version]
- Liu, C.; Goel, P.; Kaeser, P.S. Spatial and Temporal Scales of Dopamine Transmission. Nat. Rev. Neurosci. 2021, 22, 345–358. [Google Scholar] [CrossRef]
- Marcott, P.F.; Mamaligas, A.A.; Ford, C.P. Phasic Dopamine Release Drives Rapid Activation of Striatal D2-Receptors. Neuron 2014, 84, 164–176. [Google Scholar] [CrossRef] [Green Version]
- Ofori, S.; Bugnon, O.; Schorderet, M. Agonist-Induced Desensitization of Dopamine D-1 Receptors in Bovine Retina and Rat Striatum. J. Pharmacol. Exp. Ther. 1993, 266, 350–357. [Google Scholar]
- Memo, M.; Lovenberg, W.; Hanbauer, I. Agonist-Induced Subsensitivity of Adenylate Cyclase Coupled with a Dopamine Receptor in Slices from Rat Corpus Striatum. Proc. Natl. Acad. Sci. USA 1982, 79, 4456–4460. [Google Scholar] [CrossRef] [Green Version]
- Savell, K.E.; Tuscher, J.J.; Zipperly, M.E.; Duke, C.G.; Phillips, R.A.; Bauman, A.J.; Thukral, S.; Sultan, F.A.; Goska, N.A.; Ianov, L.; et al. A Dopamine-Induced Gene Expression Signature Regulates Neuronal Function and Cocaine Response. Sci. Adv. 2020, 6, eaba4221. [Google Scholar] [CrossRef]
- Scimemi, A.; Beato, M. Determining the Neurotransmitter Concentration Profile at Active Synapses. Mol. Neurobiol. 2009, 40, 289–306. [Google Scholar] [CrossRef] [Green Version]
- Berke, J.D.; Paletzki, R.F.; Aronson, G.J.; Hyman, S.E.; Gerfen, C.R. A Complex Program of Striatal Gene Expression Induced by Dopaminergic Stimulation. J. Neurosci. 1998, 18, 5301–5310. [Google Scholar] [CrossRef]
- Attisano, L.; Wrana, J.L. Signal Integration in TGF-β, WNT, and Hippo Pathways. F1000Prime Rep. 2013, 5, 17. [Google Scholar] [CrossRef]
- Weissenrieder, J.S.; Neighbors, J.D.; Mailman, R.B.; Hohl, R.J. Cancer and the Dopamine D(2) Receptor: A Pharmacological Perspective. J. Pharmacol. Exp. Ther. 2019, 370, 111–126. [Google Scholar] [CrossRef]
- Sobczuk, P.; Łomiak, M.; Cudnoch-Jędrzejewska, A. Dopamine D1 Receptor in Cancer. Cancers 2020, 12, 3232. [Google Scholar] [CrossRef]
- Zipperly, M.E.; Sultan, F.A.; Graham, G.-E.; Brane, A.C.; Simpkins, N.A.; Carullo, N.V.N.; Ianov, L.; Day, J.J. Regulation of Dopamine-Dependent Transcription and Cocaine Action by Gadd45b. Neuropsychopharmacology 2021, 46, 709–720. [Google Scholar] [CrossRef]
- Ethell, I.M.; Ethell, D.W. Matrix Metalloproteinases in Brain Development and Remodeling: Synaptic Functions and Targets. J. Neurosci. Res. 2007, 85, 2813–2823. [Google Scholar] [CrossRef] [PubMed]
- Heller, E.A.; Cates, H.M.; Peña, C.J.; Sun, H.; Shao, N.; Feng, J.; Golden, S.A.; Herman, J.P.; Walsh, J.J.; Mazei-Robison, M.; et al. Locus-Specific Epigenetic Remodeling Controls Addiction- and Depression-Related Behaviors. Nat. Neurosci. 2014, 17, 1720–1727. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vialou, V.; Maze, I.; Renthal, W.; LaPlant, Q.C.; Watts, E.L.; Mouzon, E.; Ghose, S.; Tamminga, C.A.; Nestler, E.J. Serum Response Factor Promotes Resilience to Chronic Social Stress through the Induction of ΔFosB. J. Neurosci. 2010, 30, 14585–14592. [Google Scholar] [CrossRef] [PubMed]
- Nennig, S.E.; Schank, J.R. The Role of NFkB in Drug Addiction: Beyond Inflammation. Alcohol Alcohol. 2017, 52, 172–179. [Google Scholar] [CrossRef] [PubMed]
- Cates, H.M.; Heller, E.A.; Lardner, C.K.; Purushothaman, I.; Peña, C.J.; Walker, D.M.; Cahill, M.E.; Neve, R.L.; Shen, L.; Bagot, R.C.; et al. Transcription Factor E2F3a in Nucleus Accumbens Affects Cocaine Action via Transcription and Alternative Splicing. Biol. Psychiatry 2018, 84, 167–179. [Google Scholar] [CrossRef]
- Mansvelder, H.D.; McGehee, D.S. Cellular and Synaptic Mechanisms of Nicotine Addiction. J. Neurobiol. 2002, 53, 606–617. [Google Scholar] [CrossRef]
- Greengard, P.; Allen, P.B.; Nairn, A.C. Beyond the Dopamine Receptor: The DARPP-32/Protein Phosphatase-1 Cascade. Neuron 1999, 23, 435–447. [Google Scholar] [CrossRef] [Green Version]
- Surmeier, D.J.; Ding, J.; Day, M.; Wang, Z.; Shen, W. D1 and D2 Dopamine-Receptor Modulation of Striatal Glutamatergic Signaling in Striatal Medium Spiny Neurons. Trends Neurosci. 2007, 30, 228–235. [Google Scholar] [CrossRef]
- Swapna, I.; Bondy, B.; Morikawa, H. Differential Dopamine Regulation of Ca2+ Signaling and Its Timing Dependence in the Nucleus Accumbens. Cell Rep. 2016, 15, 563–573. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.; Nagai, T.; Ahammad, R.U.; Kuroda, K.; Nakamuta, S.; Nakano, T.; Yukinawa, N.; Funahashi, Y.; Yamahashi, Y.; Amano, M.; et al. Balance between Dopamine and Adenosine Signals Regulates the PKA/Rap1 Pathway in Striatal Medium Spiny Neurons. Neurochem. Int. 2019, 122, 8–18. [Google Scholar] [CrossRef]
- Hope, B.; Kosofsky, B.; Hyman, S.E.; Nestler, E.J. Regulation of Immediate Early Gene Expression and AP-1 Binding in the Rat Nucleus Accumbens by Chronic Cocaine. Proc. Natl. Acad. Sci. USA 1992, 89, 5764–5768. [Google Scholar] [CrossRef] [Green Version]
- Valjent, E.; Pascoli, V.; Svenningsson, P.; Paul, S.; Enslen, H.; Corvol, J.-C.; Stipanovich, A.; Caboche, J.; Lombroso, P.J.; Nairn, A.C.; et al. Regulation of a Protein Phosphatase Cascade Allows Convergent Dopamine and Glutamate Signals to Activate ERK in the Striatum. Proc. Natl. Acad. Sci. USA 2005, 102, 491–496. [Google Scholar] [CrossRef] [Green Version]
- Zhang, L.; Huang, L.; Lu, K.; Liu, Y.; Tu, G.; Zhu, M.; Ying, L.; Zhao, J.; Liu, N.; Guo, F.; et al. Cocaine-Induced Synaptic Structural Modification Is Differentially Regulated by Dopamine D1 and D3 Receptors-Mediated Signaling Pathways. Addict. Biol. 2017, 22, 1842–1855. [Google Scholar] [CrossRef]
- Jerber, J.; Seaton, D.D.; Cuomo, A.S.E.; Kumasaka, N.; Haldane, J.; Steer, J.; Patel, M.; Pearce, D.; Andersson, M.; Bonder, M.J.; et al. Population-Scale Single-Cell RNA-Seq Profiling across Dopaminergic Neuron Differentiation. Nat. Genet. 2021, 53, 304–312. [Google Scholar] [CrossRef]
- Berry, B.J.; Smith, A.S.T.; Young, J.E.; Mack, D.L. Advances and Current Challenges Associated with the Use of Human Induced Pluripotent Stem Cells in Modeling Neurodegenerative Disease. Cells Tissues Organs 2018, 205, 331–349. [Google Scholar] [CrossRef]
- Umek, N.; Geršak, B.; Vintar, N.; Šoštarič, M.; Mavri, J. Dopamine Autoxidation Is Controlled by Acidic PH. Front. Mol. Neurosci. 2018, 11, 467. [Google Scholar] [CrossRef] [Green Version]
- Robertson, G.S.; Vincent, S.R.; Fibiger, H.C. D1 and D2 Dopamine Receptors Differentially Regulate C-Fos Expression in Striatonigral and Striatopallidal Neurons. Neuroscience 1992, 49, 285–296. [Google Scholar] [CrossRef]
- Perreault, M.L.; Hasbi, A.; Alijaniaram, M.; O’Dowd, B.F.; George, S.R. Reduced Striatal Dopamine D1-D2 Receptor Heteromer Expression and Behavioural Subsensitivity in Juvenile Rats. Neuroscience 2012, 225, 130–139. [Google Scholar] [CrossRef] [Green Version]
- Qian, X.; Song, H.; Ming, G. Brain Organoids: Advances, Applications and Challenges. Development 2019, 146, dev166074. [Google Scholar] [CrossRef] [Green Version]
- Lee, K.-W.; Kim, Y.; Kim, A.M.; Helmin, K.; Nairn, A.C.; Greengard, P. Cocaine-Induced Dendritic Spine Formation in D1 and D2 Dopamine Receptor-Containing Medium Spiny Neurons in Nucleus Accumbens. Proc. Natl. Acad. Sci. USA 2006, 103, 3399–3404. [Google Scholar] [CrossRef] [Green Version]
- Penrod, R.D.; Campagna, J.; Panneck, T.; Preese, L.; Lanier, L.M. The Presence of Cortical Neurons in Striatal-Cortical Co-Cultures Alters the Effects of Dopamine and BDNF on Medium Spiny Neuron Dendritic Development. Front. Cell. Neurosci. 2015, 9, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Aebersold, M.J.; Thompson-Steckel, G.; Joutang, A.; Schneider, M.; Burchert, C.; Forró, C.; Weydert, S.; Han, H.; Vörös, J. Simple and Inexpensive Paper-Based Astrocyte Co-Culture to Improve Survival of Low-Density Neuronal Networks. Front. Neurosci. 2018, 12, 94. [Google Scholar] [CrossRef]
- Delli Carri, A.; Onorati, M.; Castiglioni, V.; Faedo, A.; Camnasio, S.; Toselli, M.; Biella, G.; Cattaneo, E. Human Pluripotent Stem Cell Differentiation into Authentic Striatal Projection Neurons. Stem Cell Rev. Reports 2013, 9, 461–474. [Google Scholar] [CrossRef]
- Cruz-Santos, M.; Cardo, L.F.; Li, M. A Novel LHX6 Reporter Cell Line for Tracking Human IPSC-Derived Cortical Interneurons. Cells 2022, 11, 853. [Google Scholar] [CrossRef]
- Renthal, W.; Nestler, E. Epigenetic Mechanisms in Drug Addiction. Mol. Med. 2008, 14, 341–350. [Google Scholar] [CrossRef] [Green Version]
- Eipper-Mains, J.E.; Kiraly, D.D.; Duff, M.O.; Horowitz, M.J.; McManus, C.J.; Eipper, B.A.; Graveley, B.R.; Mains, R.E. Effects of Cocaine and Withdrawal on the Mouse Nucleus Accumbens Transcriptome. Genes, Brain Behav. 2013, 12, 21–33. [Google Scholar] [CrossRef] [PubMed] [Green Version]





Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tam, R.W.; Keung, A.J. Human Pluripotent Stem Cell-Derived Medium Spiny Neuron-like Cells Exhibit Gene Desensitization. Cells 2022, 11, 1411. https://doi.org/10.3390/cells11091411
Tam RW, Keung AJ. Human Pluripotent Stem Cell-Derived Medium Spiny Neuron-like Cells Exhibit Gene Desensitization. Cells. 2022; 11(9):1411. https://doi.org/10.3390/cells11091411
Chicago/Turabian StyleTam, Ryan W., and Albert J. Keung. 2022. "Human Pluripotent Stem Cell-Derived Medium Spiny Neuron-like Cells Exhibit Gene Desensitization" Cells 11, no. 9: 1411. https://doi.org/10.3390/cells11091411
APA StyleTam, R. W., & Keung, A. J. (2022). Human Pluripotent Stem Cell-Derived Medium Spiny Neuron-like Cells Exhibit Gene Desensitization. Cells, 11(9), 1411. https://doi.org/10.3390/cells11091411