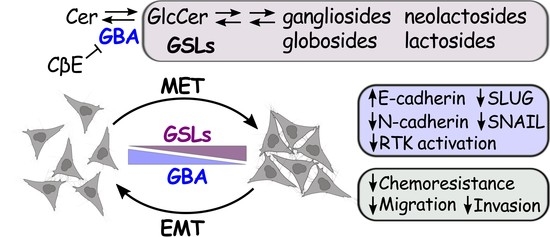
GBA Regulates EMT/MET and Chemoresistance in Squamous Cell Carcinoma Cells by Modulating the Cellular Glycosphingolipid Profile
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. Cell Viability and Proliferation Assays
2.3. Immunoblotting
2.4. Mass Spectroscopy
2.5. Untargeted Lipidomic Mass Spectrometry Analysis
2.6. GBA Knockout Cells
2.7. Statistical Analyses
2.8. Phospho-RTK Proteome Profiler Analysis
2.9. Migration Assays
2.10. Invasion Assays
2.11. Immunofluorescence Staining and Confocal Imaging
3. Results
3.1. GBA Genomic and Proteomic Alterations Are Common in Human Cancers
3.2. Depletion of GBA in HeLa and H1703 Cells Resulted in GlcCer Accumulation and Alters Their EMT/MET State
3.3. The GBA Inhibitor CβE Did Not Affect EMT Status
3.4. GBA Depletion Reduces the Migratory and Invasive Capacity of SCC Cells
3.5. GBA-Depleted SCC Cells Are Sensitized to Chemotherapeutic Agents
3.6. GBA Depletion Results in Broad Changes Receptor Tyrosine Kinase Activation
3.7. GBA Depletion Remodels the Cellular GSL Profile
4. Discussion
4.1. Differential Effect of GBA Depletion in Various Cancer Cell Lines
4.2. Role of GBA on Chemoresistance
4.3. Link among GBA Activity, RTKs, and EMT
4.4. Limitations of the Study
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhuo, D.; Li, X.; Guan, F. Biological roles of aberrantly expressed glycosphingolipids and related enzymes in human cancer development and progression. Front. Physiol. 2018, 9, 466. [Google Scholar] [CrossRef] [PubMed]
- Julien, S.; Bobowski, M.; Steenackers, A.; Le Bourhis, X.; Delannoy, P. How do gangliosides regulate RTKs signaling? Cells 2013, 2, 751–767. [Google Scholar] [CrossRef] [Green Version]
- Kaucic, K.; Liu, Y.; Ladisch, S. Modulation of growth factor signaling by gangliosides: Positive or negative? Methods Enzymol. 2006, 417, 168–185. [Google Scholar] [CrossRef] [Green Version]
- Levade, T.; Andrieu-Abadie, N.; Micheau, O.; Legembre, P.; Ségui, B. Sphingolipids modulate the epithelial–mesenchymal transition in cancer. Cell Death Discov. 2015, 1, 15001. [Google Scholar] [CrossRef] [PubMed]
- Cumin, C.; Huang, Y.-L.; Everest-Dass, A.; Jacob, F. Deciphering the importance of glycosphingolipids on cellular and molecular mechanisms associated with epithelial-to-mesenchymal transition in cancer. Biomolecules 2021, 11, 62. [Google Scholar] [CrossRef] [PubMed]
- Cumin, C.; Huang, Y.L.; Rossdam, C.; Ruoff, F.; Céspedes, S.P.; Liang, C.Y.; Lombardo, F.C.; Coelho, R.; Rimmer, N.; Konantz, M.; et al. Glycosphingolipids are mediators of cancer plasticity through independent signaling pathways. Cell Rep. 2022, 40, 111181. [Google Scholar] [CrossRef]
- Morad, S.A.F.; Cabot, M.C. Chapter Nine—The Onus of Sphingolipid Enzymes in Cancer Drug Resistance. In Advances in Cancer Research; Chalfant, C.E., Fisher, P.B., Eds.; Academic Press: New York, NY, USA, 2018; Volume 140, pp. 235–263. [Google Scholar]
- Boyd, A.E.; Grizzard, P.J.; Hylton Rorie, K.; Lima, S. Lipidomic profiling reveals biological differences between tumors of self-identified african americans and non-hispanic whites with cancer. Cancers 2023, 15, 2238. [Google Scholar] [CrossRef]
- Rohrbach, T.D.; Boyd, A.E.; Grizzard, P.J.; Spiegel, S.; Allegood, J.; Lima, S. A simple method for sphingolipid analysis of tissues embedded in optimal cutting temperature compound. J. Lipid Res. 2020, 61, 953–967. [Google Scholar] [CrossRef]
- Boyd, A.E.; Allegood, J.; Lima, S. Preparation of human tissues embedded in optimal cutting temperature compound for mass spectrometry analysis. J. Vis. Exp. 2021, 170, e62552. [Google Scholar] [CrossRef]
- Sandhoff, K.; Kolter, T. Biosynthesis and degradation of mammalian glycosphingolipids. Philos. Trans. R. Soc. Lond. Ser. B Biol. Sci. 2003, 358, 847–861. [Google Scholar] [CrossRef]
- Janneh, A.H.; Ogretmen, B. Targeting sphingolipid metabolism as a therapeutic strategy in cancer treatment. Cancers 2022, 14, 2183. [Google Scholar] [CrossRef] [PubMed]
- Ogretmen, B.; Hannun, Y.A. Biologically active sphingolipids in cancer pathogenesis and treatment. Nat. Rev. Cancer 2004, 4, 604–616. [Google Scholar] [CrossRef] [PubMed]
- Rosenbloom, B.E.; Weinreb, N.J. Gaucher disease: A comprehensive review. Crit. Rev. Oncog. 2013, 18, 163–175. [Google Scholar] [CrossRef] [PubMed]
- Gornati, R.; Berra, B.; Montorfano, G.; Martini, C.; Ciana, G.; Ferrari, P.; Romano, M.; Bembi, B. Glycolipid analysis of different tissues and cerebrospinal fluid in type II Gaucher disease. J. Inherit. Metab. Dis. 2002, 25, 47–55. [Google Scholar] [CrossRef]
- Hein, L.K.; Duplock, S.; Hopwood, J.J.; Fuller, M. Lipid composition of microdomains is altered in a cell model of Gaucher disease. J. Lipid Res. 2008, 49, 1725–1734. [Google Scholar] [CrossRef] [Green Version]
- Bodennec, J.; Pelled, D.; Riebeling, C.; Trajkovic, S.; Futerman, A.H. Phosphatidylcholine synthesis is elevated in neuronal models of Gaucher disease due to direct activation of CTP: Phosphocholine cytidylyltransferase by glucosylceramide. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 2002, 16, 1814–1816. [Google Scholar] [CrossRef]
- Batta, G.; Soltész, L.; Kovács, T.; Bozó, T.; Mészár, Z.; Kellermayer, M.; Szöllősi, J.; Nagy, P. Alterations in the properties of the cell membrane due to glycosphingolipid accumulation in a model of Gaucher disease. Sci. Rep. 2018, 8, 157. [Google Scholar] [CrossRef] [Green Version]
- Sidransky, E.; Nalls, M.A.; Aasly, J.O.; Aharon-Peretz, J.; Annesi, G.; Barbosa, E.R.; Bar-Shira, A.; Berg, D.; Bras, J.; Brice, A.; et al. Multicenter analysis of glucocerebrosidase mutations in Parkinson’s disease. N. Engl. J. Med. 2009, 361, 1651–1661. [Google Scholar] [CrossRef] [Green Version]
- Smith, L.; Schapira, A.H.V. GBA variants and Parkinson disease: Mechanisms and treatments. Cells 2022, 11, 1261. [Google Scholar] [CrossRef]
- Fuller, M. Sphingolipids: The nexus between Gaucher disease and insulin resistance. Lipids Health Dis. 2010, 9, 113. [Google Scholar] [CrossRef] [Green Version]
- Qiu, Z.; Wang, X.; Yang, Z.; Liao, S.; Dong, W.; Sun, T.; Wu, H.; Zhang, Q.; Pan, Z.; Lam, S.M.; et al. GBA1-dependent membrane glucosylceramide reprogramming promotes liver cancer metastasis via activation of the Wnt/β-catenin signalling pathway. Cell Death Dis. 2022, 13, 508. [Google Scholar] [CrossRef]
- Li, Z.; Xu, D.; Tong, X.; Shan, C. Inhibition of β-glucosidase overcomes gastric cancer chemoresistance through inducing lysosomal dysfunction. Clin. Res. Hepatol. Gastroenterol. 2021, 45, 101456. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Ouyang, B.; Jing, F.; Dai, X. GBA inhibition suppresses ovarian cancer growth, survival and receptor tyrosine kinase AXL-mediated signaling pathways. Korean J. Physiol. Pharmacol. 2023, 27, 21–29. [Google Scholar] [CrossRef] [PubMed]
- Miljan, E.A.; Bremer, E.G. Regulation of growth factor receptors by gangliosides. Sci. STKE Signal Transduct. Knowl. Environ. 2002, 2002, re15. [Google Scholar] [CrossRef]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef] [Green Version]
- Fu, J.; Zhang, Y.; Wang, Y.; Zhang, H.; Liu, J.; Tang, J.; Yang, Q.; Sun, H.; Qiu, W.; Ma, Y.; et al. Optimization of metabolomic data processing using NOREVA. Nat. Protoc. 2022, 17, 129–151. [Google Scholar] [CrossRef] [PubMed]
- Shao, X.; Lv, N.; Liao, J.; Long, J.; Xue, R.; Ai, N.; Xu, D.; Fan, X. Copy number variation is highly correlated with differential gene expression: A pan-cancer study. BMC Med. Genet. 2019, 20, 175. [Google Scholar] [CrossRef] [PubMed]
- Uhlen, M.; Zhang, C.; Lee, S.; Sjöstedt, E.; Fagerberg, L.; Bidkhori, G.; Benfeitas, R.; Arif, M.; Liu, Z.; Edfors, F.; et al. A pathology atlas of the human cancer transcriptome. Science 2017, 357, eaan2507. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bartha, Á.; Győrffy, B. TNMplot.com: A web tool for the comparison of gene expression in normal, tumor and metastatic tissues. Int. J. Mol. Sci. 2021, 22, 2622. [Google Scholar] [CrossRef]
- Kanehisa, M.; Furumichi, M.; Sato, Y.; Kawashima, M.; Ishiguro-Watanabe, M. KEGG for taxonomy-based analysis of pathways and genomes. Nucleic Acids Res. 2023, 51, D587–D592. [Google Scholar] [CrossRef]
- Grossman, R.L.; Heath, A.P.; Ferretti, V.; Varmus, H.E.; Lowy, D.R.; Kibbe, W.A.; Staudt, L.M. Toward a shared vision for cancer genomic data. New Engl. J. Med. 2016, 375, 1109–1112. [Google Scholar] [CrossRef]
- Loh, C.Y.; Chai, J.Y.; Tang, T.F.; Wong, W.F.; Sethi, G.; Shanmugam, M.K.; Chong, P.P.; Looi, C.Y. The e-cadherin and n-cadherin switch in epithelial-to-mesenchymal transition: Signaling, therapeutic implications, and challenges. Cells 2019, 8, 1118. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, J.; Antin, P.; Berx, G.; Blanpain, C.; Brabletz, T.; Bronner, M.; Campbell, K.; Cano, A.; Casanova, J.; Christofori, G.; et al. Guidelines and definitions for research on epithelial–mesenchymal transition. Nat. Rev. Mol. Cell Biol. 2020, 21, 341–352. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kuburich, N.A.; den Hollander, P.; Pietz, J.T.; Mani, S.A. Vimentin and cytokeratin: Good alone, bad together. Semin. Cancer Biol. 2022, 86, 816–826. [Google Scholar] [CrossRef]
- Tian, Y.; Qi, P.; Niu, Q.; Hu, X. Combined snail and e-cadherin predicts overall survival of cervical carcinoma patients: Comparison among various epithelial-mesenchymal transition proteins. Front. Mol. Biosci. 2020, 7, 22. [Google Scholar] [CrossRef]
- Riechelmann, H.; Steinbichler, T.B.; Sprung, S.; Santer, M.; Runge, A.; Ganswindt, U.; Gamerith, G.; Dudas, J. The epithelial-mesenchymal transcription factor slug predicts survival benefit of up-front surgery in head and neck cancer. Cancers 2021, 13, 772. [Google Scholar] [CrossRef]
- Hasan, M.R.; Sharma, R.; Saraya, A.; Chattopadhyay, T.K.; DattaGupta, S.; Walfish, P.G.; Chauhan, S.S.; Ralhan, R. Slug is a predictor of poor prognosis in esophageal squamous cell carcinoma patients. PLoS ONE 2013, 8, e82846. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shih, J.-Y.; Yang, P.-C. The EMT regulator slug and lung carcinogenesis. Carcinogenesis 2011, 32, 1299–1304. [Google Scholar] [CrossRef] [Green Version]
- Chen, B.; Chen, B.; Zhu, Z.; Ye, W.; Zeng, J.; Liu, G.; Wang, S.; Gao, J.; Xu, G.; Huang, Z. Prognostic value of ZEB-1 in solid tumors: A meta-analysis. BMC Cancer 2019, 19, 635. [Google Scholar] [CrossRef]
- Shayman, J.A. Eliglustat tartrate: Glucosylceramide synthase inhibitor treatment of type 1 Gaucher disease. Drugs Future 2010, 35, 613–620. [Google Scholar] [CrossRef]
- Van Roy, F.; Berx, G. The cell-cell adhesion molecule e-cadherin. Cell. Mol. Life Sci. CMLS 2008, 65, 3756–3788. [Google Scholar] [CrossRef]
- Zheng, X.; Carstens, J.L.; Kim, J.; Scheible, M.; Kaye, J.; Sugimoto, H.; Wu, C.C.; LeBleu, V.S.; Kalluri, R. Epithelial-to-mesenchymal transition is dispensable for metastasis but induces chemoresistance in pancreatic cancer. Nature 2015, 527, 525–530. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fischer, K.R.; Durrans, A.; Lee, S.; Sheng, J.; Li, F.; Wong, S.T.; Choi, H.; El Rayes, T.; Ryu, S.; Troeger, J.; et al. Epithelial-to-mesenchymal transition is not required for lung metastasis but contributes to chemoresistance. Nature 2015, 527, 472–476. [Google Scholar] [CrossRef] [Green Version]
- De Las Rivas, J.; Brozovic, A.; Izraely, S.; Casas-Pais, A.; Witz, I.P.; Figueroa, A. Cancer drug resistance induced by EMT: Novel therapeutic strategies. Arch. Toxicol. 2021, 95, 2279–2297. [Google Scholar] [CrossRef] [PubMed]
- Peinado, H.; Olmeda, D.; Cano, A. Snail, Zeb and bHLH factors in tumour progression: An alliance against the epithelial phenotype? Nat. Rev. Cancer 2007, 7, 415–428. [Google Scholar] [CrossRef] [PubMed]
- Corcoran, C.; O’Driscoll, L. Receptor tyrosine kinases and drug resistance: Development and characterization of in vitro models of resistance to RTK inhibitors. Methods Mol. Biol. 2015, 1233, 169–180. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.Y.; Lee, S.Y.; Yun, S.H.; Jeong, J.W.; Kim, J.H.; Kim, H.W.; Choi, J.S.; Kim, G.D.; Joo, S.T.; Choi, I.; et al. Review of the current research on fetal bovine serum and the development of cultured meat. Food Sci. Anim. Resour. 2022, 42, 775–799. [Google Scholar] [CrossRef]
- Gouaze, V.; Yu, J.Y.; Bleicher, R.J.; Han, T.Y.; Liu, Y.Y.; Wang, H.; Gottesman, M.M.; Bitterman, A.; Giuliano, A.E.; Cabot, M.C. Overexpression of glucosylceramide synthase and p-glycoprotein in cancer cells selected for resistance to natural product chemotherapy. Mol. Cancer Ther. 2004, 3, 633–639. [Google Scholar] [CrossRef]
- Gouazé, V.; Liu, Y.Y.; Prickett, C.S.; Yu, J.Y.; Giuliano, A.E.; Cabot, M.C. Glucosylceramide synthase blockade down-regulates p-glycoprotein and resensitizes multidrug-resistant breast cancer cells to anticancer drugs. Cancer Res. 2005, 65, 3861–3867. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.; Li, J.; Qiu, Z.; Gao, P.; Wu, X.; Zhou, G. Co-suppression of MDR1 (multidrug resistance 1) and GCS (glucosylceramide synthase) restores sensitivity to multidrug resistance breast cancer cells by RNA interference (RNAi). Cancer Biol. Ther. 2009, 8, 1117–1121. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.-Y.; Gupta, V.; Patwardhan, G.A.; Bhinge, K.; Zhao, Y.; Bao, J.; Mehendale, H.; Cabot, M.C.; Li, Y.-T.; Jazwinski, S.M. Glucosylceramide synthase upregulates MDR1 expression in the regulation of cancer drug resistance through cSrc and β-catenin signaling. Mol. Cancer 2010, 9, 145. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eckford, P.D.; Sharom, F.J. The reconstituted P-glycoprotein multidrug transporter is a flippase for glucosylceramide and other simple glycosphingolipids. Biochem. J. 2005, 389, 517–526. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dudas, J.; Ladanyi, A.; Ingruber, J.; Steinbichler, T.B.; Riechelmann, H. Epithelial to mesenchymal transition: A mechanism that fuels cancer radio/chemoresistance. Cells 2020, 9, 428. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shibue, T.; Weinberg, R.A. EMT, CSCs, and drug resistance: The mechanistic link and clinical implications. Nat. Rev. Clin. Oncol. 2017, 14, 611–629. [Google Scholar] [CrossRef] [Green Version]
- Kuo, C.-L.; Kallemeijn, W.W.; Lelieveld, L.T.; Mirzaian, M.; Zoutendijk, I.; Vardi, A.; Futerman, A.H.; Meijer, A.H.; Spaink, H.P.; Overkleeft, H.S.; et al. In vivo inactivation of glycosidases by conduritol B epoxide and cyclophellitol as revealed by activity-based protein profiling. FEBS J. 2019, 286, 584–600. [Google Scholar] [CrossRef] [Green Version]
- Zhuo, D.; Guan, F. Ganglioside GM1 promotes contact inhibition of growth by regulating the localization of epidermal growth factor receptor from glycosphingolipid-enriched microdomain to caveolae. Cell Prolif. 2019, 52, e12639. [Google Scholar] [CrossRef] [Green Version]
- Coskun, Ü.; Grzybek, M.; Drechsel, D.; Simons, K. Regulation of human EGF receptor by lipids. Proc. Natl. Acad. Sci. USA 2011, 108, 9044–9048. [Google Scholar] [CrossRef]
- Hein, L.K.; Meikle, P.J.; Hopwood, J.J.; Fuller, M. Secondary sphingolipid accumulation in a macrophage model of Gaucher disease. Mol. Genet. Metab. 2007, 92, 336–345. [Google Scholar] [CrossRef]
- Edmond, V.; Dufour, F.; Poiroux, G.; Shoji, K.; Malleter, M.; Fouqué, A.; Tauzin, S.; Rimokh, R.; Sergent, O.; Penna, A.; et al. Downregulation of ceramide synthase-6 during epithelial-to-mesenchymal transition reduces plasma membrane fluidity and cancer cell motility. Oncogene 2015, 34, 996–1005. [Google Scholar] [CrossRef]
- Krejci, P.; Aklian, A.; Kaucka, M.; Sevcikova, E.; Prochazkova, J.; Masek, J.K.; Mikolka, P.; Pospisilova, T.; Spoustova, T.; Weis, M.; et al. Receptor tyrosine kinases activate canonical WNT/β-catenin signaling via MAP kinase/LRP6 pathway and direct β-catenin phosphorylation. PLoS ONE 2012, 7, e35826. [Google Scholar] [CrossRef]
- Graham, T.R.; Zhau, H.E.; Odero-Marah, V.A.; Osunkoya, A.O.; Kimbro, K.S.; Tighiouart, M.; Liu, T.; Simons, J.W.; O’Regan, R.M. Insulin-like growth factor-I-dependent up-regulation of ZEB1 drives epithelial-to-mesenchymal transition in human prostate cancer cells. Cancer Res. 2008, 68, 2479–2488. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rodriguez-Monterrosas, C.; Díaz-Aragon, R.; Leal-Orta, E.; Cortes-Reynosa, P.; Perez Salazar, E. Insulin induces an EMT-like process in mammary epithelial cells MCF10A. J. Cell. Biochem. 2018, 119, 4061–4071. [Google Scholar] [CrossRef] [PubMed]
- Heldin, C.-H. Targeting the PDGF signaling pathway in tumor treatment. Cell Commun. Signal. 2013, 11, 97. [Google Scholar] [CrossRef] [Green Version]
- Antony, J.; Huang, R.Y. AXL-driven EMT state as a targetable conduit in cancer. Cancer Res. 2017, 77, 3725–3732. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, J.; Liu, S.; Yang, X.; Cao, S.; Zhou, Y. Paracrine HGF promotes EMT and mediates the effects of PSC on chemoresistance by activating c-Met/PI3K/Akt signaling in pancreatic cancer in vitro. Life Sci. 2020, 263, 118523. [Google Scholar] [CrossRef] [PubMed]
- Lo, H.W.; Hsu, S.C.; Xia, W.; Cao, X.; Shih, J.Y.; Wei, Y.; Abbruzzese, J.L.; Hortobagyi, G.N.; Hung, M.C. Epidermal growth factor receptor cooperates with signal transducer and activator of transcription 3 to induce epithelial-mesenchymal transition in cancer cells via up-regulation of TWIST gene expression. Cancer Res. 2007, 67, 9066–9076. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Langeveld, M.; Aerts, J.M.F.G. Glycosphingolipids and insulin resistance. Prog. Lipid Res. 2009, 48, 196–205. [Google Scholar] [CrossRef]
- Wang, Y.; Shi, J.; Chai, K.; Ying, X.; Zhou, B.P. The role of snail in EMT and tumorigenesis. Curr. Cancer Drug Targets 2013, 13, 963–972. [Google Scholar] [CrossRef] [Green Version]









Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Clark, L.E.; Dickinson, A.J.G.; Lima, S. GBA Regulates EMT/MET and Chemoresistance in Squamous Cell Carcinoma Cells by Modulating the Cellular Glycosphingolipid Profile. Cells 2023, 12, 1886. https://doi.org/10.3390/cells12141886
Clark LE, Dickinson AJG, Lima S. GBA Regulates EMT/MET and Chemoresistance in Squamous Cell Carcinoma Cells by Modulating the Cellular Glycosphingolipid Profile. Cells. 2023; 12(14):1886. https://doi.org/10.3390/cells12141886
Chicago/Turabian StyleClark, Laura E., Amanda J. G. Dickinson, and Santiago Lima. 2023. "GBA Regulates EMT/MET and Chemoresistance in Squamous Cell Carcinoma Cells by Modulating the Cellular Glycosphingolipid Profile" Cells 12, no. 14: 1886. https://doi.org/10.3390/cells12141886
APA StyleClark, L. E., Dickinson, A. J. G., & Lima, S. (2023). GBA Regulates EMT/MET and Chemoresistance in Squamous Cell Carcinoma Cells by Modulating the Cellular Glycosphingolipid Profile. Cells, 12(14), 1886. https://doi.org/10.3390/cells12141886