Functional Characterisation of the Circular RNA, circHTT(2-6), in Huntington’s Disease
Abstract
:1. Introduction
2. Materials and Methods
2.1. Human Ethics Approval and Patient Tissue Samples
2.2. Mouse Major Organ and Brain Tissue Harvesting
2.3. RNA and DNA Purification from Tissue
2.4. Cell Culture
2.5. Generation of circRNA Overexpression Constructs
2.6. RT-PCR/qRT-PCR
2.7. Western Blotting
2.8. Proliferation Assay
2.9. High-Content Cell Morphology Analysis
2.10. Immunofluorescence
2.11. Huntington’s Disease Diagnostic PCR
3. Results
3.1. CircRNAs from the Huntingtin (HTT) Gene
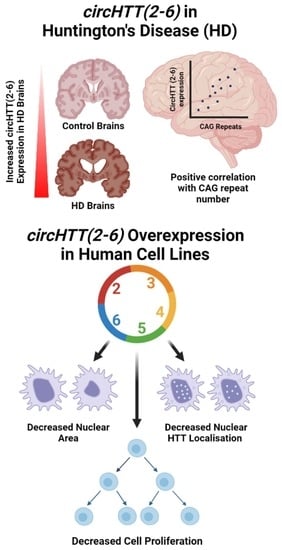
3.2. circHTT(2-6) Is a Bona Fide circRNA Enriched in HD Patient Brains
3.3. Expression Profiling of Mouse Orthologue, mmucircHTT(2-6)
3.4. Overexpression of circHTT(2-6)
3.5. HD Diagnostic PCR
3.6. Cell Morphology Analysis
3.7. Cell Proliferation
3.8. Cell Cycle Analysis by Flow Cytometry
3.9. HTT Protein Quantification and Subcellular Localisation
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sánchez, A.; Milà, M.; Castellví-Bel, S.; Rosich, M.; Jiménez, D.; Badenas, C.; Estivill, X. Maternal Transmission in Sporadic Huntington’s Disease. J. Neurol. Neurosurg. Psychiatry 1997, 62, 535–537. [Google Scholar] [CrossRef]
- The Huntington’s Disease Collaborative Research Group A Novel Gene Containing a Trinucleotide Repeat That Is Expanded and Unstable on Huntington’s Disease Chromosomes. Cell 1993, 72, 971–983. [CrossRef] [PubMed]
- Saudou, F.; Humbert, S. The Biology of Huntingtin. Neuron 2016, 89, 910–926. [Google Scholar] [CrossRef] [PubMed]
- Nasir, J.; Floresco, S.B.; O’Kusky, J.R.; Diewert, V.M.; Richman, J.M.; Zeisler, J.; Borowski, A.; Marth, J.D.; Phillips, A.G.; Hayden, M.R. Targeted Disruption of the Huntington’s Disease Gene Results in Embryonic Lethality and Behavioral and Morphological Changes in Heterozygotes. Cell 1995, 81, 811–823. [Google Scholar] [CrossRef] [PubMed]
- Zeitlin, S.; Liu, J.P.; Chapman, D.L.; Papaioannou, V.E.; Efstratiadis, A. Increased Apoptosis and Early Embryonic Lethality in Mice Nullizygous for the Huntington’s Disease Gene Homologue. Nat. Genet. 1995, 11, 155–163. [Google Scholar] [CrossRef]
- Vonsattel, J.P.; DiFiglia, M. Huntington Disease. J. Neuropathol. Exp. Neurol. 1998, 57, 369–384. [Google Scholar] [CrossRef]
- Bunner, K.D.; Rebec, G.V. Corticostriatal Dysfunction in Huntington’s Disease: The Basics. Front. Hum. Neurosci. 2016, 10, 317. [Google Scholar] [CrossRef]
- Wang, E.T.; Sandberg, R.; Luo, S.; Khrebtukova, I.; Zhang, L.; Mayr, C.; Kingsmore, S.F.; Schroth, G.P.; Burge, C.B. Alternative Isoform Regulation in Human Tissue Transcriptomes. Nature 2008, 456, 470–476. [Google Scholar] [CrossRef]
- Sibley, C.R.; Blazquez, L.; Ule, J. Lessons from Non-Canonical Splicing. Nat. Rev. Genet. 2016, 17, 407–421. [Google Scholar] [CrossRef]
- Yeo, G.; Holste, D.; Kreiman, G.; Burge, C.B. Variation in Alternative Splicing across Human Tissues. Genome Biol. 2004, 5, R74. [Google Scholar] [CrossRef]
- Scotti, M.M.; Swanson, M.S. RNA Mis-Splicing in Disease. Nat. Rev. Genet. 2016, 17, 19–32. [Google Scholar] [CrossRef]
- Jeck, W.R.; Sorrentino, J.A.; Wang, K.; Slevin, M.K.; Burd, C.E.; Liu, J.; Marzluff, W.F.; Sharpless, N.E. Circular RNAs Are Abundant, Conserved, and Associated with ALU Repeats. RNA 2013, 19, 141–157. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Wilusz, J.E.; Chen, L.-L. Biogenesis and Regulatory Roles of Circular RNAs. Annu. Rev. Cell Dev. Biol. 2022, 38, 263–289. [Google Scholar] [CrossRef] [PubMed]
- Ashwal-Fluss, R.; Meyer, M.; Pamudurti, N.R.; Ivanov, A.; Bartok, O.; Hanan, M.; Evantal, N.; Memczak, S.; Rajewsky, N.; Kadener, S. CircRNA Biogenesis Competes with Pre-MRNA Splicing. Mol. Cell 2014, 56, 55–66. [Google Scholar] [CrossRef] [PubMed]
- Conn, S.; Pillman, K.; Toubia, J.; Conn, V.; Salmanidis, M.; Phillips, C.; Roslan, S.; Schreiber, A.; Gregory, P.; Goodall, G. The RNA Binding Protein Quaking Regulates Formation of CircRNAs. Cell 2015, 160, 1125–1134. [Google Scholar] [CrossRef]
- Lukiw, W.J. Circular RNA (CircRNA) in Alzheimer’s Disease (AD). Front. Genet. 2013, 4, 307. [Google Scholar] [CrossRef]
- Hansen, T.B.; Jensen, T.I.; Clausen, B.H.; Bramsen, J.B.; Finsen, B.; Damgaard, C.K.; Kjems, J. Natural RNA Circles Function as Efficient MicroRNA Sponges. Nature 2013, 495, 384–388. [Google Scholar] [CrossRef]
- Conn, V.M.; Gabryelska, M.; Marri, S.; Stringer, B.W.; Ormsby, R.J.; Penn, T.; Poonnoose, S.; Kichenadasse, G.; Conn, S.J. SRRM4 Expands the Repertoire of Circular RNAs by Regulating Microexon Inclusion. Cells 2020, 9, 2488. [Google Scholar] [CrossRef]
- Conn, V.; Conn, S.J. SplintQuant: A Method for Accurately Quantifying Circular RNA Transcript Abundance without Reverse Transcription Bias. RNA 2019, 25, 1202–1210. [Google Scholar] [CrossRef]
- Kelley, J.B.; Paschal, B.M. Fluorescence-Based Quantification of Nucleocytoplasmic Transport. Methods 2019, 157, 106–114. [Google Scholar] [CrossRef]
- Bañez-Coronel, M.; Porta, S.; Kagerbauer, B.; Mateu-Huertas, E.; Pantano, L.; Ferrer, I.; Guzmán, M.; Estivill, X.; Martí, E. A Pathogenic Mechanism in Huntington’s Disease Involves Small CAG-Repeated RNAs with Neurotoxic Activity. PLoS Genet. 2012, 8, e1002481. [Google Scholar] [CrossRef] [PubMed]
- Slanzi, A.; Iannoto, G.; Rossi, B.; Zenaro, E.; Constantin, G. In Vitro Models of Neurodegenerative Diseases. Front. Cell Dev. Biol. 2020, 8, 328. [Google Scholar] [CrossRef] [PubMed]
- Conn, V.M.; Hugouvieux, V.; Nayak, A.; Conos, S.A.; Capovilla, G.; Cildir, G.; Jourdain, A.; Tergaonkar, V.; Schmid, M.; Zubieta, C.; et al. A CircRNA from SEPALLATA3 Regulates Splicing of Its Cognate MRNA through R-Loop Formation. Nat. Plants 2017, 3, 17053. [Google Scholar] [CrossRef] [PubMed]
- de Castro, I.J.; Toner, B.; Xie, S.Q.; Swingland, J.; Hodges, A.; Tabrizi, S.J.; Turkheimer, F.; Pombo, A.; Khalil, A. Altered Nuclear Architecture in Blood Cells from Huntington’s Disease Patients. Neurol. Sci. 2022, 43, 379–385. [Google Scholar] [CrossRef]
- Gharaba, S.; Paz, O.; Feld, L.; Abashidze, A.; Weinrab, M.; Muchtar, N.; Baransi, A.; Shalem, A.; Sprecher, U.; Wolf, L.; et al. Perturbed Actin Cap as a New Personalized Biomarker in Primary Fibroblasts of Huntington’s Disease Patients. Front. Cell Dev. Biol. 2023, 11, 1013721. [Google Scholar] [CrossRef]
- Grima, J.C.; Daigle, J.G.; Arbez, N.; Cunningham, K.C.; Zhang, K.; Ochaba, J.; Geater, C.; Morozko, E.; Stocksdale, J.; Glatzer, J.C.; et al. Mutant Huntingtin Disrupts the Nuclear Pore Complex. Neuron 2017, 94, 93–107. [Google Scholar] [CrossRef]
- Chen, X.; Zhou, M.; Yant, L.; Huang, C. Circular RNA in Disease: Basic Properties and Biomedical Relevance. Wiley Interdiscip. Rev. RNA 2022, 13, e1723. [Google Scholar] [CrossRef]
- Gasparini, S.; Licursi, V.; Presutti, C.; Mannironi, C. The Secret Garden of Neuronal CircRNAs. Cells 2020, 9, 1815. [Google Scholar] [CrossRef]
- Rybak-Wolf, A.; Stottmeister, C.; Glažar, P.; Jens, M.; Pino, N.; Giusti, S.; Hanan, M.; Behm, M.; Bartok, O.; Ashwal-Fluss, R.; et al. Circular RNAs in the Mammalian Brain Are Highly Abundant, Conserved, and Dynamically Expressed. Mol. Cell 2015, 58, 870–885. [Google Scholar] [CrossRef]
- Calderon-Villalon, J.; Ramirez-Garcia, G.; Fernandez-Ruiz, J.; Sangri-Gil, F.; Campos-Romo, A.; Galvez, V. Planning Deficits in Huntington’s Disease: A Brain Structural Correlation by Voxel-Based Morphometry. PLoS ONE 2021, 16, e0249144. [Google Scholar] [CrossRef]
- Cicchetti, F.; Prensa, L.; Wu, Y.; Parent, A. Chemical Anatomy of Striatal Interneurons in Normal Individuals and in Patients with Huntington’s Disease. Brain Res. Brain Res. Rev. 2000, 34, 80–101. [Google Scholar] [CrossRef] [PubMed]
- Orozco-Díaz, R.; Sánchez-Álvarez, A.; Hernández-Hernández, J.M.; Tapia-Ramírez, J. The Interaction between RE1-Silencing Transcription Factor (REST) and Heat Shock Protein 90 as New Therapeutic Target against Huntington’s Disease. PLoS ONE 2019, 14, e0220393. [Google Scholar] [CrossRef] [PubMed]
- Wright, G.E.B.; Collins, J.A.; Kay, C.; McDonald, C.; Dolzhenko, E.; Xia, Q.; Bečanović, K.; Drögemöller, B.I.; Semaka, A.; Nguyen, C.M.; et al. Length of Uninterrupted CAG, Independent of Polyglutamine Size, Results in Increased Somatic Instability, Hastening Onset of Huntington Disease. Am. J. Hum. Genet. 2019, 104, 1116–1126. [Google Scholar] [CrossRef]
- Jędrak, P.; Mozolewski, P.; Węgrzyn, G.; Więckowski, M.R. Mitochondrial Alterations Accompanied by Oxidative Stress Conditions in Skin Fibroblasts of Huntington’s Disease Patients. Metab. Brain Dis. 2018, 33, 2005–2017. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Lou, Y.; Cui, J.-C.; Chen, Y.; Liu, J.-T.; Yuan, Y.; Han, Y.; Huo, Y.-L.; Qi, Y.-X.; Jiang, Z.-L.; et al. Circular RNA UVRAG Mediated by Alternative Splicing Factor NOVA1 Regulates Adhesion and Migration of Vascular Smooth Muscle Cells. Genes 2021, 12, 418. [Google Scholar] [CrossRef]
- Tajima, Y.; Ito, K.; Yuan, Y.; Frank, M.O.; Saito, Y.; Darnell, R.B. NOVA1 Acts on Impact to Regulate Hypothalamic Function and Translation in Inhibitory Neurons. Cell Rep. 2023, 42, 112050. [Google Scholar] [CrossRef]
- Vedam-Mai, V.; Gardner, B.; Okun, M.S.; Siebzehnrubl, F.A.; Kam, M.; Aponso, P.; Steindler, D.A.; Yachnis, A.T.; Neal, D.; Oliver, B.U.; et al. Increased Precursor Cell Proliferation after Deep Brain Stimulation for Parkinson’s Disease: A Human Study. PLoS ONE 2014, 9, e88770. [Google Scholar] [CrossRef]
- Jin, K.; Peel, A.L.; Mao, X.O.; Xie, L.; Cottrell, B.A.; Henshall, D.C.; Greenberg, D.A. Increased Hippocampal Neurogenesis in Alzheimer’s Disease. Proc. Natl. Acad. Sci. USA 2004, 101, 343–347. [Google Scholar] [CrossRef]
- Rodríguez, J.J.; Jones, V.C.; Verkhratsky, A. Impaired Cell Proliferation in the Subventricular Zone in an Alzheimer’s Disease Model. Neuroreport 2009, 20, 907–912. [Google Scholar] [CrossRef]
- Curtis, M.A.; Penney, E.B.; Pearson, A.G.; van Roon-Mom, W.M.C.; Butterworth, N.J.; Dragunow, M.; Connor, B.; Faull, R.L.M. Increased Cell Proliferation and Neurogenesis in the Adult Human Huntington’s Disease Brain. Proc. Natl. Acad. Sci. USA 2003, 100, 9023–9027. [Google Scholar] [CrossRef]
- Askeland, G.; Dosoudilova, Z.; Rodinova, M.; Klempir, J.; Liskova, I.; Kuśnierczyk, A.; Bjørås, M.; Nesse, G.; Klungland, A.; Hansikova, H.; et al. Increased Nuclear DNA Damage Precedes Mitochondrial Dysfunction in Peripheral Blood Mononuclear Cells from Huntington’s Disease Patients. Sci. Rep. 2018, 8, 9817. [Google Scholar] [CrossRef] [PubMed]
- Wu, D.-P.; Zhao, Y.-D.; Yan, Q.-Q.; Liu, L.-L.; Wei, Y.-S.; Huang, J.-L. Circular RNAs: Emerging Players in Brain Aging and Neurodegenerative Diseases. J. Pathol. 2023, 259, 1–9. [Google Scholar] [CrossRef]
- Gasset-Rosa, F.; Chillon-Marinas, C.; Goginashvili, A.; Atwal, R.S.; Artates, J.W.; Tabet, R.; Wheeler, V.C.; Bang, A.G.; Cleveland, D.W.; Lagier-Tourenne, C. Polyglutamine-Expanded Huntingtin Exacerbates Age-Related Disruption of Nuclear Integrity and Nucleocytoplasmic Transport. Neuron 2017, 94, 48–57. [Google Scholar] [CrossRef] [PubMed]
- Sari, Y. Huntington’s Disease: From Mutant Huntingtin Protein to Neurotrophic Factor Therapy. Int. J. Biomed. Sci. IJBS 2011, 7, 89–100. [Google Scholar] [PubMed]
- Riguet, N.; Mahul-Mellier, A.-L.; Maharjan, N.; Burtscher, J.; Croisier, M.; Knott, G.; Hastings, J.; Patin, A.; Reiterer, V.; Farhan, H.; et al. Nuclear and Cytoplasmic Huntingtin Inclusions Exhibit Distinct Biochemical Composition, Interactome and Ultrastructural Properties. Nat. Commun. 2021, 12, 6579. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Zhang, X.; Li, C.; Yue, L.; Ding, N.; Riordan, T.; Yang, L.; Li, Y.; Jen, C.; Lin, S.; et al. Circular RNA Profiling Provides Insights into Their Subcellular Distribution and Molecular Characteristics in HepG2 Cells. RNA Biol. 2019, 16, 220–232. [Google Scholar] [CrossRef]
- Didiot, M.-C.; Ferguson, C.M.; Ly, S.; Coles, A.H.; Smith, A.O.; Bicknell, A.A.; Hall, L.M.; Sapp, E.; Echeverria, D.; Pai, A.A.; et al. Nuclear Localization of Huntingtin MRNA Is Specific to Cells of Neuronal Origin. Cell Rep. 2018, 24, 2553–2560.e5. [Google Scholar] [CrossRef]
- Zuccato, C.; Tartari, M.; Crotti, A.; Goffredo, D.; Valenza, M.; Conti, L.; Cataudella, T.; Leavitt, B.R.; Hayden, M.R.; Timmusk, T.; et al. Huntingtin Interacts with REST/NRSF to Modulate the Transcription of NRSE-Controlled Neuronal Genes. Nat. Genet. 2003, 35, 76–83. [Google Scholar] [CrossRef]
- Zuccato, C.; Ciammola, A.; Rigamonti, D.; Leavitt, B.R.; Goffredo, D.; Conti, L.; MacDonald, M.E.; Friedlander, R.M.; Silani, V.; Hayden, M.R.; et al. Loss of Huntingtin-Mediated BDNF Gene Transcription in Huntington’s Disease. Science 2001, 293, 493–498. [Google Scholar] [CrossRef]
- Lange, J.; Wood-Kaczmar, A.; Ali, A.; Farag, S.; Ghosh, R.; Parker, J.; Casey, C.; Uno, Y.; Kunugi, A.; Ferretti, P.; et al. Mislocalization of Nucleocytoplasmic Transport Proteins in Human Huntington’s Disease PSC-Derived Striatal Neurons. Front. Cell. Neurosci. 2021, 15, 742763. [Google Scholar] [CrossRef]
- Kim, H.J.; Taylor, J.P. Lost in Transportation: Nucleocytoplasmic Transport Defects in ALS and Other Neurodegenerative Diseases. Neuron 2017, 96, 285–297. [Google Scholar] [CrossRef] [PubMed]
- Alcalá-Vida, R.; Garcia-Forn, M.; Castany-Pladevall, C.; Creus-Muncunill, J.; Ito, Y.; Blanco, E.; Golbano, A.; Crespí-Vázquez, K.; Parry, A.; Slater, G.; et al. Neuron Type-Specific Increase in Lamin B1 Contributes to Nuclear Dysfunction in Huntington’s Disease. EMBO Mol. Med. 2021, 13, e12105. [Google Scholar] [CrossRef] [PubMed]





| Sample ID | Sex | Age at Death | Clinical Group (HD or Control) |
|---|---|---|---|
| SA0080 | Female | 81 | HD |
| SA0091 | Male | 71 | HD |
| SA0110 | Male | 72 | HD |
| SA0167 | Female | 63 | HD |
| SA0188 | Male | 64 | HD |
| SA0263 | Male | 65 | HD |
| SA0272 | Male | 60 | HD |
| SA0096 | Female | 73 | Control |
| SA0162 | Male | 72 | Control |
| SA0214 | Male | 64 | Control |
| SA0230 | Male | 86 | Control |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gantley, L.; Stringer, B.W.; Conn, V.M.; Ootsuka, Y.; Holds, D.; Slee, M.; Aliakbari, K.; Kirk, K.; Ormsby, R.J.; Webb, S.T.; et al. Functional Characterisation of the Circular RNA, circHTT(2-6), in Huntington’s Disease. Cells 2023, 12, 1337. https://doi.org/10.3390/cells12091337
Gantley L, Stringer BW, Conn VM, Ootsuka Y, Holds D, Slee M, Aliakbari K, Kirk K, Ormsby RJ, Webb ST, et al. Functional Characterisation of the Circular RNA, circHTT(2-6), in Huntington’s Disease. Cells. 2023; 12(9):1337. https://doi.org/10.3390/cells12091337
Chicago/Turabian StyleGantley, Laura, Brett W. Stringer, Vanessa M. Conn, Youichirou Ootsuka, Duncan Holds, Mark Slee, Kamelya Aliakbari, Kirsty Kirk, Rebecca J. Ormsby, Stuart T. Webb, and et al. 2023. "Functional Characterisation of the Circular RNA, circHTT(2-6), in Huntington’s Disease" Cells 12, no. 9: 1337. https://doi.org/10.3390/cells12091337
APA StyleGantley, L., Stringer, B. W., Conn, V. M., Ootsuka, Y., Holds, D., Slee, M., Aliakbari, K., Kirk, K., Ormsby, R. J., Webb, S. T., Hanson, A., Lin, H., Selth, L. A., & Conn, S. J. (2023). Functional Characterisation of the Circular RNA, circHTT(2-6), in Huntington’s Disease. Cells, 12(9), 1337. https://doi.org/10.3390/cells12091337