Emerging Picture of Deuterosome-Dependent Centriole Amplification in MCCs
Abstract
:1. Introduction
2. Centriole Structure and Organization
3. The Base of Cilia: Basal Body
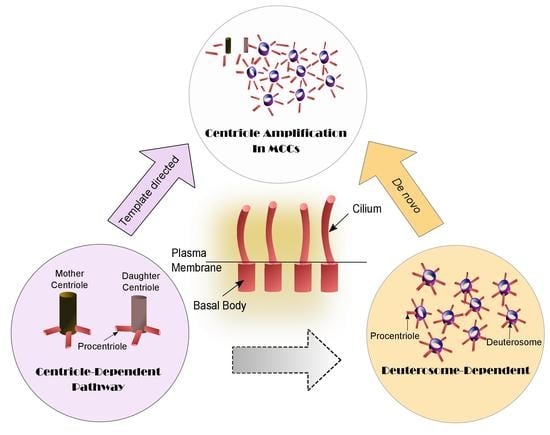
4. Centriole Amplification Pathways in Multiciliated Cells (MCCs): Centriole- and Deuterosome-Dependent Pathways
5. Changing Picture of Deuterosome-Dependent (DD) Pathway
6. Conclusions and Perspective
Acknowledgments
Conflicts of Interest
References
- Wilson, E.B. Cell in Development and Heredity, 3rd ed.; Macmillan Company: New York, NY, USA, 1925. [Google Scholar]
- Scheer, U. Historical roots of centrosome research: Discovery of Boveri’s microscope slides in Würzburg. Philos. Trans. R. Soc. B Biol. Sci. 2014, 369. [Google Scholar] [CrossRef] [PubMed]
- Bornens, M. The Centrosome in Cells and Organisms. Science 2012, 335, 422–426. [Google Scholar] [CrossRef] [PubMed]
- Schliwa, M.; Euteneuer, U.; Gräf, R.; Ueda, M. Centrosomes, microtubules and cell migration. Biochem. Soc. Symp. 1999, 65, 223–231. [Google Scholar] [PubMed]
- Arquint, C.; Gabryjonczyk, A.-M.; Nigg, E.A. Centrosomes as signalling centres. Phil. Trans. R. Soc. Lond. B Biol. Sci. 2014, 369, 20130464. [Google Scholar] [CrossRef] [PubMed]
- Werner, S.; Pimenta-Marques, A.; Bettencourt-Dias, M. Maintaining centrosomes and cilia. J. Cell Sci. 2017, 130, 3789–3800. [Google Scholar] [CrossRef] [PubMed]
- Chavali, P.L.; Pütz, M.; Gergely, F. Small organelle, big responsibility: The role of centrosomes in development and disease. Philos. Trans. R. Soc. B Biol. Sci. 2014, 369. [Google Scholar] [CrossRef] [PubMed]
- Gönczy, P. Centrosomes and cancer: Revisiting a long-standing relationship. Nat. Rev. Cancer 2015, 15, 639–652. [Google Scholar] [CrossRef] [PubMed]
- Nigg, E.A. Centrosome aberrations: Cause or consequence of cancer progression? Nat. Rev. Cancer 2002, 2, 815–825. [Google Scholar] [CrossRef] [PubMed]
- Nano, M.; Basto, R. Consequences of Centrosome Dysfunction During Brain Development. In Cell Division Machinery and Disease; Advances in Experimental Medicine and Biology; Springer: Cham, Switzerland, 2017; pp. 19–45. ISBN 978-3-319-57125-6. [Google Scholar]
- Sieben, C.; Douglass, K.M.; Guichard, P.; Manley, S. Super-resolution microscopy to decipher multi-molecular assemblies. Curr. Opin. Struct. Biol. 2018, 49, 169–176. [Google Scholar] [CrossRef] [PubMed]
- Winey, M.; O’Toole, E. Centriole structure. Philos. Trans. R. Soc. B Biol. Sci. 2014, 369. [Google Scholar] [CrossRef] [PubMed]
- Fırat-Karalar, E.N.; Stearns, T. The centriole duplication cycle. Philos. Trans. R. Soc. B Biol. Sci. 2014, 369. [Google Scholar] [CrossRef]
- Ferree, P.M.; McDonald, K.; Fasulo, B.; Sullivan, W. The Origin of Centrosomes in Parthenogenetic Hymenopteran Insects. Curr. Biol. 2006, 16, 801–807. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Riparbelli, M.G.; Callaini, G. Drosophila parthenogenesis: A model for de novo centrosome assembly. Dev. Biol. 2003, 260, 298–313. [Google Scholar] [CrossRef]
- Dirksen, E.R. Centriole Morphogenesis in Developing Ciliated Epithelium of The Mouse Oviduct. J. Cell Biol. 1971, 51, 286–302. [Google Scholar] [CrossRef] [PubMed]
- Kalnins, V.I.; Porter, K.R. Centriole replication during ciliogenesis in the chick tracheal epithelium. Z. Zellforsch. Mikrosk. Anat. 1948 1969, 100, 1–30. [Google Scholar] [CrossRef]
- Sorokin, S.P. Reconstructions of centriole formation and ciliogenesis in mammalian lungs. J. Cell Sci. 1968, 3, 207–230. [Google Scholar] [PubMed]
- Peel, N.; Stevens, N.R.; Basto, R.; Raff, J.W. Overexpressing Centriole-Replication Proteins In Vivo Induces Centriole Overduplication and De Novo Formation. Curr. Biol. 2007, 17, 834–843. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues-Martins, A.; Riparbelli, M.; Callaini, G.; Glover, D.M.; Bettencourt-Dias, M. Revisiting the Role of the Mother Centriole in Centriole Biogenesis. Science 2007, 316, 1046–1050. [Google Scholar] [CrossRef] [PubMed]
- Khodjakov, A.; Rieder, C.L.; Sluder, G.; Cassels, G.; Sibon, O.; Wang, C.-L. De novo formation of centrosomes in vertebrate cells arrested during S phase. J. Cell Biol. 2002, 158, 1171–1181. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Terra, S.L.; English, C.N.; Hergert, P.; McEwen, B.F.; Sluder, G.; Khodjakov, A. The de novo centriole assembly pathway in HeLa cells: Cell cycle progression and centriole assembly/maturation. J. Cell Biol. 2005, 168, 713–722. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.-J.; Acehan, D.; Kao, C.-H.; Jane, W.-N.; Uryu, K.; Tsou, M.-F.B. De novo centriole formation in human cells is error-prone and does not require SAS-6 self-assembly. eLife 2015, 4, e10586. [Google Scholar] [CrossRef] [PubMed]
- Steinman, R.M. An electron microscopic study of ciliogenesis in developing epidermis and trachea in the embryo of Xenopus laevis. Am. J. Anat. 1968, 122, 19–55. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Zhu, L.; Zhu, Y.; Cao, J.; Li, S.; Huang, Q.; Xu, T.; Huang, X.; Yan, X.; Zhu, X. The Cep63 paralogue Deup1 enables massive de novo centriole biogenesis for vertebrate multiciliogenesis. Nat. Cell Biol. 2013, 15, 1434–1444. [Google Scholar] [CrossRef] [PubMed]
- Klos Dehring, D.A.; Vladar, E.K.; Werner, M.E.; Mitchell, J.W.; Hwang, P.; Mitchell, B.J. Deuterosome-Mediated Centriole Biogenesis. Dev. Cell 2013, 27, 103–112. [Google Scholar] [CrossRef] [PubMed]
- Funk, M.C.; Bera, A.N.; Menchen, T.; Kuales, G.; Thriene, K.; Lienkamp, S.S.; Dengjel, J.; Omran, H.; Frank, M.; Arnold, S.J. Cyclin O (Ccno) functions during deuterosome-mediated centriole amplification of multiciliated cells. EMBO J. 2015, 34, 1078–1089. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Al Jord, A.; Lemaître, A.-I.; Delgehyr, N.; Faucourt, M.; Spassky, N.; Meunier, A. Centriole amplification by mother and daughter centrioles differs in multiciliated cells. Nature 2014, 516, 104–107. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Fernandez, J.-J.; Marshall, W.F.; Agard, D.A. Three-dimensional structure of basal body triplet revealed by electron cryo-tomography. EMBO J. 2012, 31, 552–562. [Google Scholar] [CrossRef] [PubMed]
- Gottardo, M.; Callaini, G.; Riparbelli, M.G. The Drosophila centriole—conversion of doublets into triplets within the stem cell niche. J. Cell Sci. 2015, 128, 2437–2442. [Google Scholar] [CrossRef] [PubMed]
- Pelletier, L.; O’Toole, E.; Schwager, A.; Hyman, A.A.; Müller-Reichert, T. Centriole assembly in Caenorhabditis elegans. Nature 2006, 444, 619–623. [Google Scholar] [CrossRef] [PubMed]
- Phillips, D.M. Giant Centriole Formation in Sciara. J. Cell Biol. 1967, 33, 73–92. [Google Scholar] [CrossRef] [PubMed]
- Gottardo, M.; Callaini, G.; Riparbelli, M.G. The cilium-like region of the Drosophila spermatocyte: An emerging flagellum? J. Cell Sci. 2013, 126, 5441–5452. [Google Scholar] [CrossRef] [PubMed]
- Gartenmann, L.; Wainman, A.; Qurashi, M.; Kaufmann, R.; Schubert, S.; Raff, J.W.; Dobbie, I.M. A combined 3D-SIM/SMLM approach allows centriole proteins to be localized with a precision of ∼4–5 nm. Curr. Biol. 2017, 27, R1054–R1055. [Google Scholar] [CrossRef] [PubMed]
- Van Breugel, M.; Hirono, M.; Andreeva, A.; Yanagisawa, H.; Yamaguchi, S.; Nakazawa, Y.; Morgner, N.; Petrovich, M.; Ebong, I.-O.; Robinson, C.V.; et al. Structures of SAS-6 Suggest Its Organization in Centrioles. Science 2011, 331, 1196–1199. [Google Scholar] [CrossRef] [PubMed]
- Kitagawa, D.; Vakonakis, I.; Olieric, N.; Hilbert, M.; Keller, D.; Olieric, V.; Bortfeld, M.; Erat, M.C.; Flückiger, I.; Gönczy, P.; et al. Structural Basis of the 9-Fold Symmetry of Centrioles. Cell 2011, 144, 364–375. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Andersen, J.S.; Wilkinson, C.J.; Mayor, T.; Mortensen, P.; Nigg, E.A.; Mann, M. Proteomic characterization of the human centrosome by protein correlation profiling. Nature 2003, 426, 570–574. [Google Scholar] [CrossRef] [PubMed]
- Leidel, S.; Gönczy, P. Centrosome Duplication and Nematodes: Recent Insights from an Old Relationship. Dev. Cell 2005, 9, 317–325. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Müller, H.; Schmidt, D.; Steinbrink, S.; Mirgorodskaya, E.; Lehmann, V.; Habermann, K.; Dreher, F.; Gustavsson, N.; Kessler, T.; Lehrach, H.; et al. Proteomic and functional analysis of the mitotic Drosophila centrosome. EMBO J. 2010, 29, 3344–3357. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brown, N.J.; Marjanović, M.; Lüders, J.; Stracker, T.H.; Costanzo, V. Cep63 and Cep152 Cooperate to Ensure Centriole Duplication. PLoS ONE 2013, 8, e69986. [Google Scholar] [CrossRef]
- Sonnen, K.F.; Gabryjonczyk, A.-M.; Anselm, E.; Stierhof, Y.-D.; Nigg, E.A. Human Cep192 and Cep152 cooperate in Plk4 recruitment and centriole duplication. J. Cell Sci. 2013, 126, 3223–3233. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ohta, M.; Ashikawa, T.; Nozaki, Y.; Kozuka-Hata, H.; Goto, H.; Inagaki, M.; Oyama, M.; Kitagawa, D. Direct interaction of Plk4 with STIL ensures formation of a single procentriole per parental centriole. Nat. Commun. 2014, 5, 5267. [Google Scholar] [CrossRef] [PubMed]
- Cottee, M.A.; Muschalik, N.; Wong, Y.L.; Johnson, C.M.; Johnson, S.; Andreeva, A.; Oegema, K.; Lea, S.M.; Raff, J.W.; van Breugel, M. Crystal structures of the CPAP/STIL complex reveal its role in centriole assembly and human microcephaly. eLife 2013, 2, e01071. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hatzopoulos, G.N.; Erat, M.C.; Cutts, E.; Rogala, K.B.; Slater, L.M.; Stansfeld, P.J.; Vakonakis, I. Structural analysis of the G-box domain of the microcephaly protein CPAP suggests a role in centriole architecture. Structure 2013, 21, 2069–2077. [Google Scholar] [CrossRef] [PubMed]
- Kitagawa, D.; Kohlmaier, G.; Keller, D.; Strnad, P.; Balestra, F.R.; Flückiger, I.; Gönczy, P. Spindle positioning in human cells relies on proper centriole formation and on the microcephaly proteins CPAP and STIL. J. Cell Sci. 2011, 124, 3884–3893. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tang, C.-J.C.; Lin, S.-Y.; Hsu, W.-B.; Lin, Y.-N.; Wu, C.-T.; Lin, Y.-C.; Chang, C.-W.; Wu, K.-S.; Tang, T.K. The human microcephaly protein STIL interacts with CPAP and is required for procentriole formation. EMBO J. 2011, 30, 4790. [Google Scholar] [CrossRef] [PubMed]
- Kleylein-Sohn, J.; Westendorf, J.; Le Clech, M.; Habedanck, R.; Stierhof, Y.-D.; Nigg, E.A. Plk4-Induced Centriole Biogenesis in Human Cells. Dev. Cell 2007, 13, 190–202. [Google Scholar] [CrossRef] [PubMed]
- Leidel, S.; Delattre, M.; Cerutti, L.; Baumer, K.; Gönczy, P. SAS-6 defines a protein family required for centrosome duplication in C. elegans and in human cells. Nat. Cell Biol. 2005, 7, 115–125. [Google Scholar] [CrossRef] [PubMed]
- Arquint, C.; Sonnen, K.F.; Stierhof, Y.-D.; Nigg, E.A. Cell-cycle-regulated expression of STIL controls centriole number in human cells. J. Cell Sci. 2012, 125, 1342–1352. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vulprecht, J.; David, A.; Tibelius, A.; Castiel, A.; Konotop, G.; Liu, F.; Bestvater, F.; Raab, M.S.; Zentgraf, H.; Izraeli, S.; et al. STIL is required for centriole duplication in human cells. J. Cell Sci. 2012, 125, 1353–1362. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goetz, S.C.; Anderson, K.V. The primary cilium: A signalling centre during vertebrate development. Nat. Rev. Genet. 2010, 11, 331–344. [Google Scholar] [CrossRef] [PubMed]
- Ishikawa, H.; Marshall, W.F. Ciliogenesis: Building the cell’s antenna. Nat. Rev. Mol. Cell Biol. 2011, 12, 222–234. [Google Scholar] [CrossRef] [PubMed]
- Satir, P.; Christensen, S.T. Overview of Structure and Function of Mammalian Cilia. Annu. Rev. Physiol. 2007, 69, 377–400. [Google Scholar] [CrossRef] [PubMed]
- Nigg, E.A.; Raff, J.W. Centrioles, Centrosomes, and Cilia in Health and Disease. Cell 2009, 139, 663–678. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, T.; Dynlacht, B.D. Regulating the transition from centriole to basal body. J. Cell Biol. 2011, 193, 435–444. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gilula, N.B.; Satir, P. The ciliary necklace. A ciliary membrane specialization. J. Cell Biol. 1972, 53, 494–509. [Google Scholar] [CrossRef] [PubMed]
- Lechtreck, K.F. IFT-cargo interactions and protein transport in cilia. Trends Biochem. Sci. 2015, 40, 765–778. [Google Scholar] [CrossRef] [PubMed]
- Mottier-Pavie, V.; Megraw, T.L. Drosophila Bld10 Is a Centriolar Protein That Regulates Centriole, Basal Body, and Motile Cilium Assembly. Mol. Biol. Cell 2009, 20, 2605–2614. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tanos, B.E.; Yang, H.-J.; Soni, R.; Wang, W.-J.; Macaluso, F.P.; Asara, J.M.; Tsou, M.-F.B. Centriole distal appendages promote membrane docking, leading to cilia initiation. Genes Dev. 2013, 27, 163–168. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, N.; Xia, Y.; Zhang, D.; Wang, S.; Bao, Y.; He, R.; Teng, J.; Chen, J. Hierarchical assembly of centriole subdistal appendages via centrosome binding proteins CCDC120 and CCDC68. Nat. Commun. 2017, 8, 15057. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, J.; Gao, J.; Adamian, M.; Wen, X.-H.; Pawlyk, B.; Zhang, L.; Sanderson, M.J.; Zuo, J.; Makino, C.L.; Li, T. The Ciliary Rootlet Maintains Long-Term Stability of Sensory Cilia. Mol. Cell. Biol. 2005, 25, 4129–4137. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, J.V.; Kao, L.-R.; Jana, S.C.; Sivan-Loukianova, E.; Mendonça, S.; Cabrera, O.A.; Singh, P.; Cabernard, C.; Eberl, D.F.; Bettencourt-Dias, M.; et al. Rootletin organizes the ciliary rootlet to achieve neuron sensory function in Drosophila. J. Cell Biol. 2015, 211, 435–453. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Allen, R.D. The morphogenesis of basal bodies and accessory structures of the cortex of the ciliated protozoan Tetrahymena pyriformis. J. Cell Biol. 1969, 40, 716–733. [Google Scholar] [CrossRef] [PubMed]
- Machemer, H. Ciliary activity and the origin of metachrony in Paramecium: Effects of increased viscosity. J. Exp. Biol. 1972, 57, 239–259. [Google Scholar] [PubMed]
- Hodges, M.E.; Wickstead, B.; Gull, K.; Langdale, J.A. The evolution of land plant cilia. New Phytol. 2012, 195, 526–540. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wanner, A.; Salathé, M.; O’Riordan, T.G. Mucociliary clearance in the airways. Am. J. Respir. Crit. Care Med. 1996, 154, 1868–1902. [Google Scholar] [CrossRef] [PubMed]
- Worthington, W.C.; Cathcart, R.S. Ependymal cilia: Distribution and activity in the adult human brain. Science 1963, 139, 221–222. [Google Scholar] [CrossRef] [PubMed]
- Fliegauf, M.; Benzing, T.; Omran, H. When cilia go bad: Cilia defects and ciliopathies. Nat. Rev. Mol. Cell Biol. 2007, 8, 880–893. [Google Scholar] [CrossRef] [PubMed]
- Al Jord, A.; Shihavuddin, A.; d’Aout, R.S.; Faucourt, M.; Genovesio, A.; Karaiskou, A.; Sobczak-Thépot, J.; Spassky, N.; Meunier, A. Calibrated mitotic oscillator drives motile ciliogenesis. Science 2017, 358, 803–806. [Google Scholar] [CrossRef] [PubMed]
- Revinski, D.R.; Zaragosi, L.-E.; Boutin, C.; Ruiz-Garcia, S.; Deprez, M.; Rosnet, O.; Thome, V.; Mercey, O.; Paquet, A.; Pons, N.; et al. CDC20B is required for deuterosome-mediated centriole production in multiciliated cells. bioRxiv 2017, 218750. [Google Scholar] [CrossRef] [Green Version]
- Anderson, R.G.; Brenner, R.M. The formation of basal bodies (centrioles) in the Rhesus monkey oviduct. J. Cell Biol. 1971, 50, 10–34. [Google Scholar] [CrossRef] [PubMed]
- Baccetti, B.; Dallai, R. The spermatozoon of arthropoda. XXX. The multiflagellate spermatozoon in the termite Mastotermes darwiniensis. J. Cell Biol. 1978, 76, 569–576. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Buckland-Nicks, J. Prosobranch parasperm: Sterile germ cells that promote paternity? Micron 1998, 29, 267–280. [Google Scholar] [CrossRef]
- Riparbelli, M.G.; Stouthamer, R.; Dallai, R.; Callaini, G. Microtubule organization during the early development of the parthenogenetic egg of the hymenopteran Muscidifurax uniraptor. Dev. Biol. 1998, 195, 89–99. [Google Scholar] [CrossRef] [PubMed]
- Marescalchi, O.; Zauli, C.; Scali, V. Centrosome dynamics and inheritance in related sexual and parthenogenetic Bacillus (Insecta Phasmatodea). Mol. Reprod. Dev. 2002, 63, 89–95. [Google Scholar] [CrossRef] [PubMed]
- Heath, I.B.; Kaminskyj, S.G.; Bauchop, T. Basal body loss during fungal zoospore encystment: Evidence against centriole autonomy. J. Cell Sci. 1986, 83, 135–140. [Google Scholar] [PubMed]
- Mizukami, I.; Gall, J. CENTRIOLE REPLICATION: II. Sperm Formation in the Fern, Marsilea, and the Cycad, Zamia. J. Cell Biol. 1966, 29, 97–111. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Saint Phalle, B.; Sullivan, W. Spindle assembly and mitosis without centrosomes in parthenogenetic Sciara embryos. J. Cell Biol. 1998, 141, 1383–1391. [Google Scholar] [CrossRef] [PubMed]
- Fulton, C.; Dingle, A.D. Basal Bodies, but Not Centrioles, in Naegleria. J. Cell Biol. 1971, 51, 826–836. [Google Scholar] [CrossRef] [PubMed]
- Grimes, G.W. Morphological Discontinuity of Kinetosomes During the Life Cycle of Oxytricha Fallax. J. Cell Biol. 1973, 57, 229–232. [Google Scholar] [CrossRef] [PubMed]
- Sluder, G.; Miller, F.J.; Rieder, C.L. Reproductive capacity of sea urchin centrosomes without centrioles. Cell Motil. Cytoskeleton 1989, 13, 264–273. [Google Scholar] [CrossRef] [PubMed]
- Szöllosi, D.; Ozil, J.P. De novo formation of centrioles in parthenogenetically activated, diploidized rabbit embryos. Biol. Cell 1991, 72, 61–66. [Google Scholar] [CrossRef]
- Vladar, E.K.; Stearns, T. Molecular characterization of centriole assembly in ciliated epithelial cells. J. Cell Biol. 2007, 178, 31–42. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mori, M.; Hazan, R.; Danielian, P.S.; Mahoney, J.E.; Li, H.; Lu, J.; Miller, E.S.; Zhu, X.; Lees, J.A.; Cardoso, W.V. Cytoplasmic E2f4 forms organizing centres for initiation of centriole amplification during multiciliogenesis. Nat. Commun. 2017, 8, 15857. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, S.; Ma, L.; Shokhirev, M.N.; Quigley, I.; Kintner, C. Multicilin and activated E2f4 induce multiciliated cell differentiation in primary fibroblasts. Sci. Rep. 2018, 8, 12369. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Chen, Q.; Huang, Q.; Yan, X.; Zhu, X. Mother centrioles are dispensable for deuterosome formation and function during basal body amplification. bioRxiv 2018, 373662. [Google Scholar] [CrossRef]
- Ma, L.; Quigley, I.; Omran, H.; Kintner, C. Multicilin drives centriole biogenesis via E2f proteins. Genes Dev. 2014, 28, 1461–1471. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boon, M.; Wallmeier, J.; Ma, L.; Loges, N.T.; Jaspers, M.; Olbrich, H.; Dougherty, G.W.; Raidt, J.; Werner, C.; Amirav, I.; et al. MCIDAS mutations result in a mucociliary clearance disorder with reduced generation of multiple motile cilia. Nat. Commun. 2014, 5, 4418. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Terré, B.; Piergiovanni, G.; Segura-Bayona, S.; Gil-Gómez, G.; Youssef, S.A.; Attolini, C.S.-O.; Wilsch-Bräuninger, M.; Jung, C.; Rojas, A.M.; Marjanović, M.; et al. GEMC1 is a critical regulator of multiciliated cell differentiation. EMBO J. 2016, 35, 942–960. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, F.; Narasimhan, V.; Shboul, M.; Chong, Y.L.; Reversade, B.; Roy, S. Gmnc Is a Master Regulator of the Multiciliated Cell Differentiation Program. Curr. Biol. 2015, 25, 3267–3273. [Google Scholar] [CrossRef] [PubMed]


© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shahid, U.; Singh, P. Emerging Picture of Deuterosome-Dependent Centriole Amplification in MCCs. Cells 2018, 7, 152. https://doi.org/10.3390/cells7100152
Shahid U, Singh P. Emerging Picture of Deuterosome-Dependent Centriole Amplification in MCCs. Cells. 2018; 7(10):152. https://doi.org/10.3390/cells7100152
Chicago/Turabian StyleShahid, Umama, and Priyanka Singh. 2018. "Emerging Picture of Deuterosome-Dependent Centriole Amplification in MCCs" Cells 7, no. 10: 152. https://doi.org/10.3390/cells7100152
APA StyleShahid, U., & Singh, P. (2018). Emerging Picture of Deuterosome-Dependent Centriole Amplification in MCCs. Cells, 7(10), 152. https://doi.org/10.3390/cells7100152