New Insights into the Runt Domain of RUNX2 in Melanoma Cell Proliferation and Migration
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bioinformatic Investigation of RUNX2 and Associated Genes in Melanoma
2.2. Cell Cultures
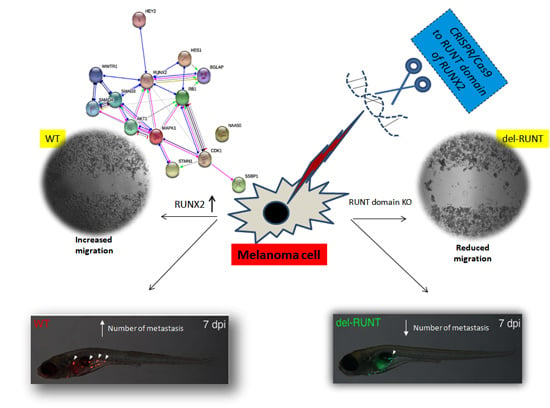
2.3. CRISPR/Cas9-Mediated Deletion of the RUNT Domain from RUNX2
2.4. XTT Test
2.5. Cell Proliferation Test
2.6. TUNEL Test
2.7. RUNX2 Transfection
2.8. Total RNA Extraction and Reverse Transcription (RT)
2.9. Real Time RT-PCR
2.10. Migration Assay
2.11. Western Blot
2.12. Zebrafish Handling and Xenotransplantation
2.13. Statistical Analysis
3. Results
3.1. Bioinformatic Analysis of RUNX2 using the Cancer Genome Atlas (TCGA) Skin Cutaneous Melanoma Dataset
3.2. Generation of del-RUNT Cells
3.3. The RUNT Domain Increases Cell Viability and Proliferation and Reduces Apoptosis
3.4. RUNT Modulation of Genes Involved in Metastatic Progression
3.5. RUNT Domain Promotes Epithelial Mesenchymal Transition and Migration Ability
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Whiteman, D.C.; Pavan, W.J.; Bastian, B.C. The melanomas: A synthesis of epidemiological, clinical, histopathological, genetic, and biological aspects, supporting distinct subtypes, causal pathways, and cells of origin. Pigment Cell Melanoma Res. 2011, 24, 879–897. [Google Scholar] [CrossRef] [PubMed]
- Damsky, W.E.; Bosenberg, M. Melanocytic nevi and melanoma: Unraveling a complex relationship. Oncogene 2017, 36, 5771–5792. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; He, R.; Xia, H.; Wei, Y.; Wu, S. Knockdown of STMN1 enhances osteosarcoma cell chemosensitivity through inhibition of autophagy. Oncol. Lett. 2017, 13, 3465–3470. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, S.; Wu, Y.; Chen, Q.; Cao, J.; Hu, K.; Tang, J.; Sang, Y.; Lai, F.; Wang, L.; Zhang, R.; et al. hSSB1 regulates both the stability and the transcriptional activity of p53. Cell Res. 2013, 23, 423–435. [Google Scholar] [CrossRef] [PubMed]
- Cecconi, D.; Carbonare, L.D.; Mori, A.; Cheri, S.; Deiana, M.; Brandi, J.; Degaetano, V.; Masiero, V.; Innamorati, G.; Mottes, M.; et al. An integrated approach identifies new oncotargets in melanoma. Oncotarget 2018, 9, 11489–11502. [Google Scholar] [CrossRef] [PubMed]
- Timar, J.; Vizkeleti, L.; Doma, V.; Barbai, T.; Raso, E. Genetic progression of malignant melanoma. Cancer Metastasis Rev. 2016, 35, 93–107. [Google Scholar] [CrossRef] [PubMed]
- Ozturk Sari, S.; Yilmaz, I.; Taskin, O.C.; Narli, G.; Sen, F.; Comoglu, S.; Firat, P.; Bİlgİç, B.; Yilmazbayhan, D.; Ozluk, Y.; et al. BRAF, NRAS, KIT, TERT, GNAQ/GNA11 mutation profile analysis of head and neck mucosal melanomas: A study of 42 cases. Pathology 2017, 49, 55–61. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Cheng, H.S.; Chng, W.J.; Tergaonkar, V. Activation of mutant TERT promoter by RAS-ERK signaling is a key step in malignant progression of BRAF-mutant human melanomas. Proc. Natl. Acad. Sci.USA 2016, 113, 14402–14407. [Google Scholar] [CrossRef] [PubMed]
- Siroy, A.E.; Boland, G.M.; Milton, D.R.; Roszik, J.; Frankian, S.; Malke, J.; Haydu, L.; Prieto, V.G.; Tetzlaff, M.; Ivan, D.; et al. Beyond BRAF(V600): Clinical mutation panel testing by next-generation sequencing in advanced melanoma. J. Investig. Dermatol. 2015, 135, 508–515. [Google Scholar] [CrossRef] [PubMed]
- Motzik, A.; Amir, E.; Erlich, T.; Wang, J.; Kim, B.G.; Han, J.M.; Kim, J.H.; Nechushtan, H.; Guo, M.; Razin, E.; et al. Post-translational modification of HINT1 mediates activation of MITF transcriptional activity in human melanoma cells. Oncogene 2017, 36, 4732–4738. [Google Scholar] [CrossRef] [PubMed]
- Cohen-Solal, K.A.; Boregowda, R.K.; Lasfar, A. RUNX2 and the PI3K/AKT axis reciprocal activation as a driving force for tumor progression. Mol. Cancer 2015, 14. [Google Scholar] [CrossRef] [PubMed]
- Choe, M.; Brusgard, J.L.; Chumsri, S.; Bhandary, L.; Zhao, X.F.; Lu, S.; Goloubeva, O.G.; Polster, B.M.; Fiskum, G.M.; Girnun, G.D.; et al. The RUNX2 Transcription Factor Negatively Regulates SIRT6 Expression to Alter Glucose Metabolism in Breast Cancer Cells. J. Cell. Biochem. 2015, 116, 2210–2226. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ozaki, T.; Nakamura, M.; Shimozato, O. Novel Implications of DNA Damage Response in Drug Resistance of Malignant Cancers Obtained from the Functional Interaction between p53 Family and RUNX2. Biomolecules 2015, 5, 2854–2876. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Perduca, M.; Dalle Carbonare, L.; Bovi, M.; Innamorati, G.; Cheri, S.; Cavallini, C.; Scupoli, M.T.; Mori, A.; Valenti, M.T. Runx2 downregulation, migration and proliferation inhibition in melanoma cells treated with BEL beta-trefoil. Oncol. Rep. 2017, 37, 2209–2214. [Google Scholar] [CrossRef] [PubMed]
- Boregowda, R.K.; Olabisi, O.O.; Abushahba, W.; Jeong, B.S.; Haenssen, K.K.; Chen, W.; Chekmareva, M.; Lasfar, A.; Foran, D.J.; Goydos, J.S.; et al. RUNX2 is overexpressed in melanoma cells and mediates their migration and invasion. Cancer Lett. 2014, 348, 61–70. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dalle Carbonare, L.; Innamorati, G.; Valenti, M.T. Transcription factor Runx2 and its application to bone tissue engineering. Stem Cell Rev. 2012, 8, 891–897. [Google Scholar] [CrossRef] [PubMed]
- Valenti, M.T.; Serafini, P.; Innamorati, G.; Gili, A.; Cheri, S.; Bassi, C.; Carbonare, L.D. Runx2 expression: A mesenchymal stem marker for cancer. Oncol. Lett. 2016, 12, 4167–4172. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dalle Carbonare, L.; Frigo, A.; Francia, G.; Davi, M.V.; Donatelli, L.; Stranieri, C.; Brazzarola, P.; Zatelli, M.C.; Menestrina, F.; Valenti, M.T. Runx2 mRNA expression in the tissue, serum, and circulating non-hematopoietic cells of patients with thyroid cancer. J. Clin. Endocrinol. Metabol. 2012, 97, E1249–E1256. [Google Scholar] [CrossRef] [PubMed]
- Riminucci, M.; Corsi, A.; Peris, K.; Fisher, L.W.; Chimenti, S.; Bianco, P. Coexpression of bone sialoprotein (BSP) and the pivotal transcriptional regulator of osteogenesis, Cbfa1/Runx2, in malignant melanoma. Calcif. Tissue Int. 2003, 73, 281–289. [Google Scholar] [CrossRef] [PubMed]
- Cerami, E.; Gao, J.; Dogrusoz, U.; Gross, B.E.; Sumer, S.O.; Aksoy, B.A.; Jacobsen, A.; Byrne, C.J.; Heuer, M.L.; Larsson, E.; et al. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012, 2, 401–404. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Labun, K.; Montague, T.G.; Gagnon, J.A.; Thyme, S.B.; Valen, E. CHOPCHOP v2: A web tool for the next generation of CRISPR genome engineering. Nucleic Acids Res. 2016, 44, W272–W276. [Google Scholar] [CrossRef] [PubMed]
- Montague, T.G.; Cruz, J.M.; Gagnon, J.A.; Church, G.M.; Valen, E. CHOPCHOP: A CRISPR/Cas9 and TALEN web tool for genome editing. Nucleic Acids Res. 2014, 42, W401–W407. [Google Scholar] [CrossRef] [PubMed]
- Ran, F.A.; Hsu, P.D.; Wright, J.; Agarwala, V.; Scott, D.A.; Zhang, F. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 2013, 8, 2281–2308. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Valenti, M.T.; Zanatta, M.; Donatelli, L.; Viviano, G.; Cavallini, C.; Scupoli, M.T.; Carbonare, L.D. Ascorbic acid induces either differentiation or apoptosis in MG-63 osteosarcoma lineage. Anticancer Res. 2014, 34, 1617–1628. [Google Scholar] [PubMed]
- Dalle Carbonare, L.; Valenti, M.T.; Azzarello, G.; Balducci, E.; Crepaldi, G.; Realdi, G.; Vinante, O.; Giannini, S. Bisphosphonates decrease telomerase activity and hTERT expression in MCF-7 breast cancer cells. Mol Cell Endocrinol. 2005, 30, 23–31. [Google Scholar] [CrossRef] [PubMed]
- Whitlock, K.E.; Westerfield, M. The olfactory placodes of the zebrafish form by convergence of cellular fields at the edge of the neural plate. Development 2000, 127, 3645–3653. [Google Scholar] [PubMed]
- Kimmel, C.B.; Ballard, W.W.; Kimmel, S.R.; Ullmann, B.; Schilling, T.F. Stages of embryonic development of the zebrafish. Dev. Dyn. 1995, 203, 253–310. [Google Scholar] [CrossRef] [PubMed]
- Meyer, M.B.; Benkusky, N.A.; Pike, J.W. Selective Distal Enhancer Control of the Mmp13 Gene Identified through Clustered Regularly Interspaced Short Palindromic Repeat (CRISPR) Genomic Deletions. J. Biol. Chem. 2015, 290, 11093–11107. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaufman, C.K. Zebrafish Melanoma. Adv. Exp. Med. Biol. 2016, 916, 439–450. [Google Scholar] [PubMed]
- Kaufman, C.K.; Mosimann, C.; Fan, Z.P.; Yang, S.; Thomas, A.J.; Ablain, J.; Tan, J.L.; Fogley, R.D.; van Rooijen, E.; Hagedorn, E.J.; et al. A zebrafish melanoma model reveals emergence of neural crest identity during melanoma initiation. Science 2016, 351. [Google Scholar] [CrossRef] [PubMed]
- Lei, Q.Y.; Zhang, H.; Zhao, B.; Zha, Z.Y.; Bai, F.; Pei, X.H.; Zhao, S.; Xiong, Y.; Guan, K.L. TAZ promotes cell proliferation and epithelial-mesenchymal transition and is inhibited by the hippo pathway. Mol. Cell. Biol. 2008, 28, 2426–2436. [Google Scholar] [CrossRef] [PubMed]
- Shin, M.H.; He, Y.; Marrogi, E.; Piperdi, S.; Ren, L.; Khanna, C.; Gorlick, R.; Liu, C.; Huang, J. A RUNX2-Mediated Epigenetic Regulation of the Survival of p53 Defective Cancer Cells. PLoS Genet. 2016, 12. [Google Scholar] [CrossRef] [PubMed]
- Ozaki, T.; Nakagawara, A.; Nagase, H. A novel role of RUNX2 in the regulation of p53-dependent DNA damage response. Seikagaku 2013, 85, 1002–1007. [Google Scholar] [PubMed]
- Lin, W.; Zhu, X.; Yang, S.; Chen, X.; Wang, L.; Huang, Z.; Ding, Y.; Huang, L.; Lv, C. MicroRNA-203 inhibits proliferation and invasion, and promotes apoptosis of osteosarcoma cells by targeting Runt-related transcription factor 2. Biomed. Pharmacother. 2017, 91, 1075–1084. [Google Scholar] [CrossRef] [PubMed]
- Colden, M.; Dar, A.A.; Saini, S.; Dahiya, P.V.; Shahryari, V.; Yamamura, S.; Tanaka, Y.; Stein, G.; Dahiya, R.; Majid, S.; et al. MicroRNA-466 inhibits tumor growth and bone metastasis in prostate cancer by direct regulation of osteogenic transcription factor RUNX2. Cell Death Dis. 2017, 8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, H.; Jiang, T.; Ren, K.; Li, Z.L.; Ren, J.; Wu, G.; Han, X. RUNX2 Plays An Oncogenic Role in Esophageal Carcinoma by Activating the PI3K/AKT and ERK Signaling Pathways. Cell. Physiol. Biochem. 2018, 49, 217–225. [Google Scholar] [CrossRef] [PubMed]









© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Deiana, M.; Dalle Carbonare, L.; Serena, M.; Cheri, S.; Parolini, F.; Gandini, A.; Marchetto, G.; Innamorati, G.; Manfredi, M.; Marengo, E.; et al. New Insights into the Runt Domain of RUNX2 in Melanoma Cell Proliferation and Migration. Cells 2018, 7, 220. https://doi.org/10.3390/cells7110220
Deiana M, Dalle Carbonare L, Serena M, Cheri S, Parolini F, Gandini A, Marchetto G, Innamorati G, Manfredi M, Marengo E, et al. New Insights into the Runt Domain of RUNX2 in Melanoma Cell Proliferation and Migration. Cells. 2018; 7(11):220. https://doi.org/10.3390/cells7110220
Chicago/Turabian StyleDeiana, Michela, Luca Dalle Carbonare, Michela Serena, Samuele Cheri, Francesca Parolini, Alberto Gandini, Giulia Marchetto, Giulio Innamorati, Marcello Manfredi, Emilio Marengo, and et al. 2018. "New Insights into the Runt Domain of RUNX2 in Melanoma Cell Proliferation and Migration" Cells 7, no. 11: 220. https://doi.org/10.3390/cells7110220
APA StyleDeiana, M., Dalle Carbonare, L., Serena, M., Cheri, S., Parolini, F., Gandini, A., Marchetto, G., Innamorati, G., Manfredi, M., Marengo, E., Brandi, J., Cecconi, D., Mori, A., Mina, M. M., Antoniazzi, F., Mottes, M., Tiso, N., Malerba, G., Zipeto, D., & Valenti, M. T. (2018). New Insights into the Runt Domain of RUNX2 in Melanoma Cell Proliferation and Migration. Cells, 7(11), 220. https://doi.org/10.3390/cells7110220