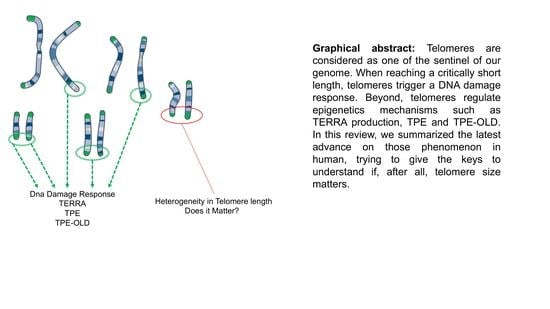
Bring It to an End: Does Telomeres Size Matter?
Abstract
:1. Introduction
2. Telomere Dynamics
2.1. Telomere Shortening
2.2. Telomere Elongation
2.3. Heritable Telomere-Length
3. Telomere Singularity: Possible Causes of Differences between Telomere Length
3.1. Telomere Nuclear Localization
3.2. Telomere Replication
4. Telomere Epigenetics and Position Effect
4.1. Epigenetic Signature
4.2. Telomere Position Effect
4.3. Telomere Position Effect-Over Long Distance
4.4. Telomeric Repeat-Containing RNA
5. Telomeropathies
6. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Muller, H.J. The remaking of chromosomes. Collect. Net 1938, 8, 182–195. [Google Scholar]
- Blackburn, E.H.; Gall, J.G. A tandemly repeated sequence at the termini of the extrachromosomal ribosomal RNA genes in Tetrahymena. J. Mol. Biol. 1978, 120, 33–53. [Google Scholar] [CrossRef]
- Greider, C.W.; Blackburn, E.H. Identification of a specific telomere terminal transferase activity in Tetrahymena extracts. Cell 1985, 43, 405–413. [Google Scholar] [CrossRef]
- Greider, C.W.; Blackburn, E.H. The telomere terminal transferase of Tetrahymena is a ribonucleoprotein enzyme with two kinds of primer specificity. Cell 1987, 51, 887–898. [Google Scholar] [CrossRef]
- Greider, C.W.; Blackburn, E.H. A telomeric sequence in the RNA of Tetrahymena telomerase required for telomere repeat synthesis. Nature 1989, 337, 331–337. [Google Scholar] [CrossRef] [PubMed]
- Moyzis, R.K.; Buckingham, J.M.; Cram, L.S.; Dani, M.; Deaven, L.L.; Jones, M.D.; Meyne, J.; Ratliff, R.L.; Wu, J.R. A highly conserved repetitive DNA sequence, (TTAGGG)n, present at the telomeres of human chromosomes. Proc. Natl. Acad. Sci. USA 1988, 85, 6622–6626. [Google Scholar] [CrossRef] [PubMed]
- Griffith, J.D.; Comeau, L.; Rosenfield, S.; Stansel, R.M.; Bianchi, A.; Moss, H.; de Lange, T. Mammalian telomeres end in a large duplex loop. Cell 1999, 97, 503–514. [Google Scholar] [CrossRef]
- Doksani, Y.; Wu, J.Y.; de Lange, T.; Zhuang, X. Super-resolution fluorescence imaging of telomeres reveals TRF2-dependent T-loop formation. Cell 2013, 155, 345–356. [Google Scholar] [CrossRef]
- Van Ly, D.; Low, R.R.J.; Frölich, S.; Bartolec, T.K.; Kafer, G.R.; Pickett, H.A.; Gaus, K.; Cesare, A.J. Telomere loop dynamics in chromosome end protection. Mol. Cell 2018, 71, 510–525. [Google Scholar] [CrossRef]
- Baumann, P. Pot1, the Putative Telomere End-Binding Protein in Fission Yeast and Humans. Science 2001, 292, 1171–1175. [Google Scholar] [CrossRef] [Green Version]
- Bilaud, T.; Brun, C.; Ancelin, K.; Koering, C.E.; Laroche, T.; Gilson, E. Telomeric localization of TRF2, a novel human telobox protein. Nat. Genet. 1997, 17, 236–239. [Google Scholar] [CrossRef] [PubMed]
- Broccoli, D.; Smogorzewska, A.; Chong, L.; de Lange, T. Human telomeres contain two distinct Myb–related proteins, TRF1 and TRF2. Nat. Genet. 1997, 17, 231–235. [Google Scholar] [CrossRef]
- Chong, L.; van Steensel, B.; Broccoli, D.; Erdjument-Bromage, H.; Hanish, J.; Tempst, P.; de Lange, T. A Human Telomeric Protein. Science 1995, 270, 1663–1667. [Google Scholar] [CrossRef] [PubMed]
- Houghtaling, B.R.; Cuttonaro, L.; Chang, W.; Smith, S. A Dynamic Molecular Link between the Telomere Length Regulator TRF1 and the Chromosome End Protector TRF2. Curr. Biol. 2004, 14, 1621–1631. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, S.; Kaminker, P.; Campisi, J. TIN2, a new regulator of telomere length in human cells. Nat. Genet. 1999, 23, 405–412. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, B.; Oestreich, S.; de Lange, T. Identification of Human Rap1. Cell 2000, 101, 471–483. [Google Scholar] [CrossRef] [Green Version]
- Liu, D.; Safari, A.; O’Connor, M.S.; Chan, D.W.; Laegeler, A.; Qin, J.; Songyang, Z. PTOP interacts with POT1 and regulates its localization to telomeres. Nat. Cell Biol. 2004, 6, 673–680. [Google Scholar] [CrossRef]
- Ye, J.Z.-S. POT1-interacting protein PIP1: A telomere length regulator that recruits POT1 to the TIN2/TRF1 complex. Genes Dev. 2004, 18, 1649–1654. [Google Scholar] [CrossRef]
- Zhong, Z.; Shiue, L.; Kaplan, S.; de Lange, T. A mammalian factor that binds telomeric TTAGGG repeats in vitro. Mol. Cell. Biol. 1992, 12, 4834–4843. [Google Scholar] [CrossRef]
- Chang, H.H.Y.; Pannunzio, N.R.; Adachi, N.; Lieber, M.R. Non-homologous DNA end joining and alternative pathways to double-strand break repair. Nat. Rev. Mol. Cell Biol. 2017, 18, 495–506. [Google Scholar] [CrossRef]
- O’Sullivan, R.J.; Karlseder, J. Telomeres: Protecting chromosomes against genome instability. Nat. Rev. Mol. Cell Biol. 2010, 11, 171–181. [Google Scholar] [CrossRef] [PubMed]
- Van Steensel, B.; Smogorzewska, A.; de Lange, T. TRF2 protects human telomeres from end-to-end fusions. Cell 1998, 92, 401–413. [Google Scholar] [CrossRef]
- Olovnikov, A.M. Principle of marginotomy in template synthesis of polynucleotides. Dokl. Akad. Nauk SSSR 1971, 201, 1496–1499. (In Russian) [Google Scholar] [PubMed]
- Watson, J.D. Origin of concatemeric T7DNA. Nat. New Biol. 1972, 239, 197–201. [Google Scholar] [CrossRef] [PubMed]
- Harley, C.B.; Futcher, A.B.; Greider, C.W. Telomeres shorten during aging of human fibroblasts. Nature 1990, 345, 458–460. [Google Scholar] [CrossRef] [PubMed]
- Ohki, R.; Tsurimoto, T.; Ishikawa, F. In vitro reconstitution of the end replication problem. Mol. Cell. Biol. 2001, 21, 5753–5766. [Google Scholar] [CrossRef]
- Palm, W.; de Lange, T. How Shelterin Protects Mammalian Telomeres. Ann. Rev. Genet. 2008, 42, 301–334. [Google Scholar] [CrossRef]
- D’Adda di Fagagna, F.; Reaper, P.M.; Clay-Farrace, L.; Fiegler, H.; Carr, P.; von Zglinicki, T.; Saretzki, G.; Carter, N.P.; Jackson, S.P. A DNA damage checkpoint response in telomere-initiated senescence. Nature 2003, 426, 194–198. [Google Scholar] [CrossRef]
- Takai, H.; Smogorzewska, A.; de Lange, T. DNA damage foci at dysfunctional telomeres. Curr. Biol. 2003, 13, 1549–1556. [Google Scholar] [CrossRef]
- Hemann, M.T.; Strong, M.A.; Hao, L.Y.; Greider, C.W. The shortest telomere, not average telomere length, is critical for cell viability and chromosome stability. Cell 2001, 107, 67–77. [Google Scholar] [CrossRef]
- Meier, A.; Fiegler, H.; Muñoz, P.; Ellis, P.; Rigler, D.; Langford, C.; Blasco, M.A.; Carter, N.; Jackson, S.P. Spreading of mammalian DNA-damage response factors studied by ChIP-chip at damaged telomeres. EMBO J. 2007, 26, 2707–2718. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zou, Y.; Sfeir, A.; Gryaznov, S.M.; Shay, J.W.; Wright, W.E. Does a sentinel or a subset of short telomeres determine replicative senescence? Mol. Biol. Cell 2004, 15, 3709–3718. [Google Scholar] [CrossRef] [PubMed]
- Daniali, L.; Benetos, A.; Susser, E.; Kark, J.D.; Labat, C.; Kimura, M.; Desai, K.K.; Granick, M.; Aviv, A. Telomeres shorten at equivalent rates in somatic tissues of adults. Nat. Commun. 2013, 4, 1597. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Montpetit, A.J.; Alhareeri, A.A.; Montpetit, M.; Starkweather, A.R.; Elmore, L.W.; Filler, K.; Mohanraj, L.; Burton, C.W.; Menzies, V.S.; Lyon, D.E.; et al. Telomere length: A review of methods for measurement. Nurs. Res. 2014, 63, 289–299. [Google Scholar] [CrossRef]
- Jaskelioff, M.; Muller, F.L.; Paik, J.-H.; Thomas, E.; Jiang, S.; Adams, A.C.; Sahin, E.; Kost-Alimova, M.; Protopopov, A.; Cadiñanos, J.; et al. Telomerase reactivation reverses tissue degeneration in aged telomerase-deficient mice. Nature 2011, 469, 102–106. [Google Scholar] [CrossRef] [PubMed]
- Bodnar, A.G.; Ouellette, M.; Frolkis, M.; Holt, S.E.; Chiu, C.P.; Morin, G.B.; Harley, C.B.; Shay, J.W.; Lichtsteiner, S.; Wright, W.E. Extension of life-span by introduction of telomerase into normal human cells. Science 1998, 279, 349–352. [Google Scholar] [CrossRef]
- Li, J.S.Z.; Miralles Fusté, J.; Simavorian, T.; Bartocci, C.; Tsai, J.; Karlseder, J.; Lazzerini Denchi, E. TZAP: A telomere-associated protein involved in telomere length control. Science 2017, 355, 638–641. [Google Scholar] [CrossRef] [PubMed]
- Holohan, B.; De Meyer, T.; Batten, K.; Mangino, M.; Hunt, S.C.; Bekaert, S.; De Buyzere, M.L.; Rietzschel, E.R.; Spector, T.D.; Wright, W.E.; et al. Decreasing initial telomere length in humans intergenerationally understates age-associated telomere shortening. Aging Cell 2015, 14, 669–677. [Google Scholar] [CrossRef] [Green Version]
- Hayashi, M.T.; Cesare, A.J.; Rivera, T.; Karlseder, J. Cell death during crisis is mediated by mitotic telomere deprotection. Nature 2015, 522, 492–496. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, N.W.; Piatyszek, M.A.; Prowse, K.R.; Harley, C.B.; West, M.D.; Ho, P.L.; Coviello, G.M.; Wright, W.E.; Weinrich, S.L.; Shay, J.W. Specific association of human telomerase activity with immortal cells and cancer. Science 1994, 266, 2011–2015. [Google Scholar] [CrossRef]
- Cong, Y.S.; Wen, J.; Bacchetti, S. The human telomerase catalytic subunit hTERT: Organization of the gene and characterization of the promoter. Hum. Mol. Genet. 1999, 8, 137–142. [Google Scholar] [CrossRef] [PubMed]
- Kim, W.; Ludlow, A.T.; Min, J.; Robin, J.D.; Stadler, G.; Mender, I.; Lai, T.-P.; Zhang, N.; Wright, W.E.; Shay, J.W. Regulation of the human telomerase gene TERT by telomere position effect—Over long distances (TPE-OLD): Implications for aging and cancer. PLoS Biol. 2016, 14, e2000016. [Google Scholar] [CrossRef] [PubMed]
- Shay, J.W.; Wright, W.E. Senescence and immortalization: Role of telomeres and telomerase. Carcinogenesis 2005, 26, 867–874. [Google Scholar] [CrossRef] [PubMed]
- Vinagre, J.; Almeida, A.; Pópulo, H.; Batista, R.; Lyra, J.; Pinto, V.; Coelho, R.; Celestino, R.; Prazeres, H.; Lima, L.; et al. Frequency of TERT promoter mutations in human cancers. Nat. Commun. 2013, 4, 2185. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- O’Sullivan, R.J.; Arnoult, N.; Lackner, D.H.; Oganesian, L.; Haggblom, C.; Corpet, A.; Almouzni, G.; Karlseder, J. Rapid induction of alternative lengthening of telomeres by depletion of the histone chaperone ASF1. Nat. Struct. Mol. Biol. 2014, 21, 167–174. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dilley, R.L.; Verma, P.; Cho, N.W.; Winters, H.D.; Wondisford, A.R.; Greenberg, R.A. Break-induced telomere synthesis underlies alternative telomere maintenance. Nature 2016, 539, 54–58. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, J.; Wang, F.; Okuka, M.; Liu, N.; Ji, G.; Ye, X.; Zuo, B.; Li, M.; Liang, P.; Ge, W.W.; et al. Association of telomere length with authentic pluripotency of ES/iPS cells. Cell Res. 2011, 21, 779–792. [Google Scholar] [CrossRef] [Green Version]
- Zalzman, M.; Falco, G.; Sharova, L.V.; Nishiyama, A.; Thomas, M.; Lee, S.-L.; Stagg, C.A.; Hoang, H.G.; Yang, H.-T.; Indig, F.E.; et al. Zscan4 regulates telomere elongation and genomic stability in ES cells. Nature 2010, 464, 858–863. [Google Scholar] [CrossRef] [Green Version]
- Zeng, S.; Liu, L.; Sun, Y.; Xie, P.; Hu, L.; Yuan, D.; Chen, D.; Ouyang, Q.; Lin, G.; Lu, G. Telomerase-mediated telomere elongation from human blastocysts to embryonic stem cells. J. Cell Sci. 2014, 127, 752–762. [Google Scholar] [CrossRef]
- Londoño-Vallejo, J.A.; DerSarkissian, H.; Cazes, L.; Thomas, G. Differences in telomere length between homologous chromosomes in humans. Nucleic Acids Res. 2001, 29, 3164–3171. [Google Scholar] [CrossRef] [Green Version]
- Graakjaer, J.; Bischoff, C.; Korsholm, L.; Holstebroe, S.; Vach, W.; Bohr, V.A.; Christensen, K.; Kølvraa, S. The pattern of chromosome-specific variations in telomere length in humans is determined by inherited, telomere-near factors and is maintained throughout life. Mech. Aging Dev. 2003, 124, 629–640. [Google Scholar] [CrossRef]
- Hjelmborg, J.B.; Dalgård, C.; Möller, S.; Steenstrup, T.; Kimura, M.; Christensen, K.; Kyvik, K.O.; Aviv, A. The heritability of leucocyte telomere length dynamics. J. Med. Genet. 2015, 52, 297–302. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fraga, M.F.; Ballestar, E.; Paz, M.F.; Ropero, S.; Setien, F.; Ballestar, M.L.; Heine-Suner, D.; Cigudosa, J.C.; Urioste, M.; Benitez, J.; et al. From the cover: Epigenetic differences arise during the lifetime of monozygotic twins. Proc. Natl. Acad. Sci. USA 2005, 102, 10604–10609. [Google Scholar] [CrossRef] [PubMed]
- Slagboom, P.E.; Droog, S.; Boomsma, D.I. Genetic determination of telomere size in humans: A twin study of three age groups. Am. J. Hum. Genet. 1994, 55, 876–882. [Google Scholar] [PubMed]
- Lansdorp, P.M.; Verwoerd, N.P.; van de Rijke, F.M.; Dragowska, V.; Little, M.T.; Dirks, R.W.; Raap, A.K.; Tanke, H.J. Heterogeneity in telomere length of human chromosomes. Hum. Mol. Genet. 1996, 5, 685–691. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martens, U.M.; Zijlmans, J.M.J.M.; Poon, S.S.S.; Dragowska, W.; Yui, J.; Chavez, E.A.; Ward, R.K.; Lansdorp, P.M. Short telomeres on human chromosome 17p. Nat. Genet. 1998, 18, 76–80. [Google Scholar] [CrossRef]
- Broer, L.; Codd, V.; Nyholt, D.R.; Deelen, J.; Mangino, M.; Willemsen, G.; Albrecht, E.; Amin, N.; Beekman, M.; de Geus, E.J.C.; et al. Meta-analysis of telomere length in 19 713 subjects reveals high heritability, stronger maternal inheritance and a paternal age effect. Eur. J. Hum. Genet. 2013, 21, 1163–1168. [Google Scholar] [CrossRef] [Green Version]
- Graakjaer, J.; Pascoe, L.; Der-Sarkissian, H.; Thomas, G.; Kolvraa, S.; Christensen, K.; Londono-Vallejo, J.-A. The relative lengths of individual telomeres are defined in the zygote and strictly maintained during life. Aging Cell 2004, 3, 97–102. [Google Scholar] [CrossRef] [Green Version]
- Guelen, L.; Pagie, L.; Brasset, E.; Meuleman, W.; Faza, M.B.; Talhout, W.; Eussen, B.H.; de Klein, A.; Wessels, L.; de Laat, W.; et al. Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature 2008, 453, 948–951. [Google Scholar] [CrossRef]
- Wang, S.; Su, J.-H.; Beliveau, B.J.; Bintu, B.; Moffitt, J.R.; Wu, C.; Zhuang, X. Spatial organization of chromatin domains and compartments in single chromosomes. Science 2016, 353, 598–602. [Google Scholar] [CrossRef] [Green Version]
- Robin, J.D.; Magdinier, F. Physiological and pathological aging affects chromatin dynamics, structure and function at the nuclear edge. Front. Genet. 2016, 7, 153. [Google Scholar] [CrossRef] [PubMed]
- Gonzalo, S.; Eissenberg, J.C. Tying up loose ends: Telomeres, genomic instability and lamins. Curr. Opin. Genet. Dev. 2016, 37, 109–118. [Google Scholar] [CrossRef] [PubMed]
- Solovei, I.; Wang, A.S.; Thanisch, K.; Schmidt, C.S.; Krebs, S.; Zwerger, M.; Cohen, T.V.; Devys, D.; Foisner, R.; Peichl, L.; et al. LBR and Lamin A/C sequentially tether peripheral heterochromatin and inversely regulate differentiation. Cell 2013, 152, 584–598. [Google Scholar] [CrossRef] [PubMed]
- Dechat, T. LAP2 and BAF transiently localize to telomeres and specific regions on chromatin during nuclear assembly. J. Cell Sci. 2004, 117, 6117–6128. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, S.; Schones, D.E.; Malicet, C.; Rochman, M.; Zhou, M.; Foisner, R.; Bustin, M. High mobility group protein N5 (HMGN5) and Lamina-associated polypeptide 2α (LAP2α) interact and reciprocally affect their genome-wide chromatin organization. J. Biol. Chem. 2013, 288, 18104–18109. [Google Scholar] [CrossRef] [PubMed]
- Wood, A.M.; Danielsen, J.M.R.; Lucas, C.A.; Rice, E.L.; Scalzo, D.; Shimi, T.; Goldman, R.D.; Smith, E.D.; Le Beau, M.M.; Kosak, S.T. TRF2 and lamin A/C interact to facilitate the functional organization of chromosome ends. Nat. Commun. 2014, 5, 5467. [Google Scholar] [CrossRef] [PubMed]
- Chojnowski, A.; Ong, P.F.; Wong, E.S.; Lim, J.S.; Mutalif, R.A.; Navasankari, R.; Dutta, B.; Yang, H.; Liow, Y.Y.; Sze, S.K.; et al. Progerin reduces LAP2α-telomere association in hutchinson-gilford progeria. ELife 2015, 4, e07759. [Google Scholar] [CrossRef] [PubMed]
- Ottaviani, A.; Schluth-Bolard, C.; Rival-Gervier, S.; Boussouar, A.; Rondier, D.; Foerster, A.M.; Morere, J.; Bauwens, S.; Gazzo, S.; Callet-Bauchu, E.; et al. Identification of a perinuclear positioning element in human subtelomeres that requires A-type lamins and CTCF. EMBO J. 2009, 28, 2428–2436. [Google Scholar] [CrossRef]
- Britt-Compton, B.; Rowson, J.; Locke, M.; Mackenzie, I.; Kipling, D.; Baird, D.M. Structural stability and chromosome-specific telomere length is governed by cis-acting determinants in humans. Hum. Mol. Genet. 2006, 15, 725–733. [Google Scholar] [CrossRef] [Green Version]
- Arnoult, N.; Schluth-Bolard, C.; Letessier, A.; Drascovic, I.; Bouarich-Bourimi, R.; Campisi, J.; Kim, S.; Boussouar, A.; Ottaviani, A.; Magdinier, F.; et al. Replication timing of human telomeres is chromosome arm–specific, influenced by subtelomeric structures and connected to nuclear localization. PLoS Genet. 2010, 6, e1000920. [Google Scholar] [CrossRef]
- Piqueret-Stephan, L.; Ricoul, M.; Hempel, W.M.; Sabatier, L. Replication timing of human telomeres is conserved during immortalization and influenced by respective subtelomeres. Sci. Rep. 2016, 6, 32510. [Google Scholar] [CrossRef] [PubMed]
- Mendez-Bermudez, A.; Lototska, L.; Bauwens, S.; Giraud-Panis, M.-J.; Croce, O.; Jamet, K.; Irizar, A.; Mowinckel, M.; Koundrioukoff, S.; Nottet, N.; et al. Genome-wide control of heterochromatin replication by the telomere capping protein TRF2. Mol. Cell 2018, 70, 449–461. [Google Scholar] [CrossRef]
- Leman, A.R.; Dheekollu, J.; Deng, Z.; Lee, S.W.; Das, M.M.; Lieberman, P.M.; Noguchi, E. Timeless preserves telomere length by promoting efficient DNA replication through human telomeres. Cell Cycle 2012, 11, 2337–2347. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamazaki, S.; Hayano, M.; Masai, H. Replication timing regulation of eukaryotic replicons: Rif1 as a global regulator of replication timing. Trends Genet. 2013, 29, 449–460. [Google Scholar] [CrossRef] [PubMed]
- Baur, J.A. Telomere Position Effect in Human Cells. Science 2001, 292, 2075–2077. [Google Scholar] [CrossRef]
- Robin, J.D.; Ludlow, A.T.; Batten, K.; Magdinier, F.; Stadler, G.; Wagner, K.R.; Shay, J.W.; Wright, W.E. Telomere position effect: Regulation of gene expression with progressive telomere shortening over long distances. Genes Dev. 2014, 28, 2464–2476. [Google Scholar] [CrossRef] [PubMed]
- Schoeftner, S.; Blasco, M.A. Developmentally regulated transcription of mammalian telomeres by DNA-dependent RNA polymerase II. Nat. Cell Biol. 2008, 10, 228–236. [Google Scholar] [CrossRef] [PubMed]
- Blasco, M.A. Telomere epigenetics: A higher-order control of telomere length in mammalian cells. Carcinogenesis 2004, 25, 1083–1087. [Google Scholar] [CrossRef]
- Cubiles, M.D.; Barroso, S.; Vaquero-Sedas, M.I.; Enguix, A.; Aguilera, A.; Vega-Palas, M.A. Epigenetic features of human telomeres. Nucleic Acids Res. 2018, 46, 2347–2355. [Google Scholar] [CrossRef] [Green Version]
- Negishi, Y.; Kawaji, H.; Minoda, A.; Usui, K. Identification of chromatin marks at TERRA promoter and encoding region. Biochem. Biophys. Res. Commun. 2015, 467, 1052–1057. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blasco, M.A. The epigenetic regulation of mammalian telomeres. Nat. Rev. Genet. 2007, 8, 299–309. [Google Scholar] [CrossRef] [PubMed]
- Creyghton, M.P.; Cheng, A.W.; Welstead, G.G.; Kooistra, T.; Carey, B.W.; Steine, E.J.; Hanna, J.; Lodato, M.A.; Frampton, G.M.; Sharp, P.A.; et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc. Natl. Acad. Sci. USA 2010, 107, 21931–21936. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gottschling, D.E.; Aparicio, O.M.; Billington, B.L.; Zakian, V.A. Position effect at S. cerevisiae telomeres: Reversible repression of Pol II transcription. Cell 1990, 63, 751–762. [Google Scholar] [CrossRef]
- Ottaviani, A.; Gilson, E.; Magdinier, F. Telomeric position effect: From the yeast paradigm to human pathologies? Biochimie 2008, 90, 93–107. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Muller, H.J. Types of visible variations induced by X-rays in Drosophila. J. Genet. 1930, 22, 299–334. [Google Scholar] [CrossRef]
- Akhtar, W.; de Jong, J.; Pindyurin, A.V.; Pagie, L.; Meuleman, W.; de Ridder, J.; Berns, A.; Wessels, L.F.A.; van Lohuizen, M.; van Steensel, B. Chromatin position effects assayed by thousands of reporters integrated in parallel. Cell 2013, 154, 914–927. [Google Scholar] [CrossRef]
- Ofir, R.; Wong, A.C.; McDermid, H.E.; Skorecki, K.L.; Selig, S. Position effect of human telomeric repeats on replication timing. Proc. Natl. Acad. Sci. USA 1999, 96, 11434–11439. [Google Scholar] [CrossRef] [Green Version]
- Koering, C.E.; Pollice, A.; Zibella, M.P.; Bauwens, S.; Puisieux, A.; Brunori, M.; Brun, C.; Martins, L.; Sabatier, L.; Pulitzer, J.F.; et al. Human telomeric position effect is determined by chromosomal context and telomeric chromatin integrity. EMBO Rep. 2002, 3, 1055–1061. [Google Scholar] [CrossRef] [Green Version]
- Kulkarni, A.; Zschenker, O.; Reynolds, G.; Miller, D.; Murnane, J.P. Effect of telomere proximity on telomere position effect, chromosome healing, and sensitivity to DNA double-strand breaks in a human tumor cell line. Mol. Cell. Biol. 2010, 30, 578–589. [Google Scholar] [CrossRef]
- Stadler, G.; Rahimov, F.; King, O.D.; Chen, J.C.J.; Robin, J.D.; Wagner, K.R.; Shay, J.W.; Emerson, C.P.; Wright, W.E. Telomere position effect regulates DUX4 in human facioscapulohumeral muscular dystrophy. Nat. Struct. Mol. Biol. 2013, 20, 671–678. [Google Scholar] [CrossRef] [Green Version]
- Tennen, R.I.; Bua, D.J.; Wright, W.E.; Chua, K.F. SIRT6 is required for maintenance of telomere position effect in human cells. Nat Commun. 2011, 2, 433. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bell, A.C.; West, A.G.; Felsenfeld, G. Insulators and boundaries: Versatile regulatory elements in the eukaryotic genome. Science 2001, 291, 447–450. [Google Scholar] [CrossRef] [PubMed]
- Recillas-Targa, F.; Pikaart, M.J.; Burgess-Beusse, B.; Bell, A.C.; Litt, M.D.; West, A.G.; Gaszner, M.; Felsenfeld, G. Position-effect protection and enhancer blocking by the chicken -globin insulator are separable activities. Proc. Natl. Acad. Sci. USA 2002, 99, 6883–6888. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rincon-Arano, H.; Furlan-Magaril, M.; Recillas-Targa, F. Protection against telomeric position effects by the chicken cHS4 beta-globin insulator. Proc. Natl. Acad. Sci. USA 2007, 104, 14044–14049. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lou, Z.; Wei, J.; Riethman, H.; Baur, J.A.; Voglauer, R.; Shay, J.W.; Wright, W.E. Telomere length regulates ISG15 expression in human cells. Aging 2009, 1, 608–621. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stone, E.M.; Pillus, L. Activation of an MAP kinase cascade leads to Sir3p hyperphosphorylation and strengthens transcriptional silencing. J. Cell Biol. 1996, 135, 571–583. [Google Scholar] [CrossRef] [Green Version]
- Ai, W.; Bertram, P.G.; Tsang, C.K.; Chan, T.F.; Zheng, X.F.S. Regulation of subtelomeric silencing during stress response. Mol. Cell 2002, 10, 1295–1305. [Google Scholar] [CrossRef]
- Halme, A.; Bumgarner, S.; Styles, C.; Fink, G.R. Genetic and epigenetic regulation of the FLO gene family generates cell-surface variation in yeast. Cell 2004, 116, 405–415. [Google Scholar] [CrossRef]
- D’Cunha, J.; Knight, E.; Haas, A.L.; Truitt, R.L.; Borden, E.C. Immunoregulatory properties of ISG15, an interferon-induced cytokine. Proc. Natl. Acad. Sci. USA 1996, 93, 211–215. [Google Scholar] [CrossRef]
- Schedl, P.; Broach, J.R. Making good neighbors: The right fence for the right job. Nat. Struct. Biol. 2003, 10, 241–243. [Google Scholar] [CrossRef]
- Mukherjee, A.K.; Sharma, S.; Sengupta, S.; Saha, D.; Kumar, P.; Hussain, T.; Srivastava, V.; Roy, S.D.; Shay, J.W.; Chowdhury, S. Telomere length-dependent transcription and epigenetic modifications in promoters remote from telomere ends. PLoS Genet. 2018, 14, e1007782. [Google Scholar] [CrossRef] [PubMed]
- Kim, W.; Shay, J.W. Long-range telomere regulation of gene expression: Telomere looping and telomere position effect over long distances (TPE-OLD). Differentiation 2018, 99, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Azzalin, C.M.; Reichenbach, P.; Khoriauli, L.; Giulotto, E.; Lingner, J. Telomeric repeat containing RNA and RNA surveillance factors at mammalian chromosome ends. Science 2007, 318, 798–801. [Google Scholar] [CrossRef] [PubMed]
- Chu, H.-P.; Cifuentes-Rojas, C.; Kesner, B.; Aeby, E.; Lee, H.; Wei, C.; Oh, H.J.; Boukhali, M.; Haas, W.; Lee, J.T. TERRA RNA antagonizes ATRX and protects telomeres. Cell 2017, 170, 86–101. [Google Scholar] [CrossRef]
- Deng, Z.; Norseen, J.; Wiedmer, A.; Riethman, H.; Lieberman, P.M. TERRA RNA binding to TRF2 facilitates heterochromatin formation and ORC recruitment at telomeres. Mol. Cell 2009, 35, 403–413. [Google Scholar] [CrossRef]
- De Silanes, I.L.; d’Alcontres, M.S.; Blasco, M.A. TERRA transcripts are bound by a complex array of RNA-binding proteins. Nat. Commun. 2010, 1, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Kupiec, M. Biology of telomeres: Lessons from budding yeast. FEMS Microbiol. Rev. 2014, 38, 144–171. [Google Scholar] [CrossRef]
- Montero, J.J.; López-Silanes, I.; Megías, D.; Fraga, M.F.; Castells-García, Á.; Blasco, M.A. TERRA recruitment of polycomb to telomeres is essential for histone trymethylation marks at telomeric heterochromatin. Nat. Commun. 2018, 9, 1548. [Google Scholar] [CrossRef]
- Diman, A.; Decottignies, A. Genomic origin and nuclear localization of TERRA telomeric repeat-containing RNA: From Darkness to Dawn. FEBS J. 2018, 285, 1389–1398. [Google Scholar] [CrossRef]
- Arora, R.; Lee, Y.; Wischnewski, H.; Brun, C.M.; Schwarz, T.; Azzalin, C.M. RNaseH1 regulates TERRA-telomeric DNA hybrids and telomere maintenance in ALT tumour cells. Nat. Commun. 2014, 5, 5220. [Google Scholar] [CrossRef] [Green Version]
- Cusanelli, E.; Romero, C.A.P.; Chartrand, P. Telomeric noncoding RNA TERRA is induced by telomere shortening to nucleate telomerase molecules at short telomeres. Mol. Cell 2013, 51, 780–791. [Google Scholar] [CrossRef] [PubMed]
- Sagie, S.; Toubiana, S.; Hartono, S.R.; Katzir, H.; Tzur-Gilat, A.; Havazelet, S.; Francastel, C.; Velasco, G.; Chédin, F.; Selig, S. Telomeres in ICF syndrome cells are vulnerable to DNA damage due to elevated DNA:RNA hybrids. Nat. Commun. 2017, 8, 14015. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bertuch, A.A. The molecular genetics of the telomere biology disorders. RNA Biol. 2016, 13, 696–706. [Google Scholar] [CrossRef] [PubMed]
- Opresko, P.L.; Shay, J.W. Telomere-associated aging disorders. Ageing Res. Rev. 2017, 33, 52–66. [Google Scholar] [CrossRef] [PubMed]
- Holohan, B.; Wright, W.E.; Shay, J.W. Telomeropathies: An emerging spectrum disorder. J. Cell Biol. 2014, 205, 289–299. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Armanios, M.Y.; Chen, J.J.-L.; Cogan, J.D.; Alder, J.K.; Ingersoll, R.G.; Markin, C.; Lawson, W.E.; Xie, M.; Vulto, I.; Phillips, J.A.; et al. Telomerase mutations in families with idiopathic pulmonary fibrosis. N. Engl. J. Med. 2007, 356, 1317–1326. [Google Scholar] [CrossRef] [PubMed]
- Heiss, N.S.; Knight, S.W.; Vulliamy, T.J.; Klauck, S.M.; Wiemann, S.; Mason, P.J.; Poustka, A.; Dokal, I. X-linked dyskeratosis congenita is caused by mutations in a highly conserved gene with putative nucleolar functions. Nat. Genet. 1998, 19, 32–38. [Google Scholar] [CrossRef] [PubMed]
- Savage, S.A. Human telomeres and telomere biology disorders. In Progress in Molecular Biology and Translational Science; Elsevier: Bethesda, MA, USA, 2014; Volume 125, pp. 41–66. ISBN 978-0-12-397898-1. [Google Scholar]
- Knight, S.J.; Regan, R.; Nicod, A.; Horsley, S.W.; Kearney, L.; Homfray, T.; Winter, R.M.; Bolton, P.; Flint, J. Subtle chromosomal rearrangements in children with unexplained mental retardation. Lancet 1999, 354, 1676–1681. [Google Scholar] [CrossRef]
- Fernandez, B.; Siegel-Bartelt, J.; Herbrick, J.-A.; Teshima, I.; Scherer, S. Holoprosencephaly and cleidocranial dysplasia in a patient due to two position-effect mutations: Case report and review of the literature: 6;7 Translocation and position-effect mutations. Clin. Genet. 2005, 68, 349–359. [Google Scholar] [CrossRef] [PubMed]
- Guilherme, R.S.; Moysés-Oliveira, M.; Dantas, A.G.; Meloni, V.A.; Colovati, M.E.; Kulikowski, L.D.; Melaragno, M.I. Position effect modifying gene expression in a patient with ring chromosome 14. J. Appl. Genet. 2016, 57, 183–187. [Google Scholar] [CrossRef]
- Surace, C.; Berardinelli, F.; Masotti, A.; Roberti, M.; Da Sacco, L.; D’Elia, G.; Sirleto, P.; Digilio, M.; Cusmai, R.; Grotta, S.; et al. Telomere shortening and telomere position effect in mild ring 17 syndrome. Epigenet. Chromatin 2014, 7, 1. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zou, Y.S.; Van Dyke, D.L.; Thorland, E.C.; Chhabra, H.S.; Michels, V.V.; Keefe, J.G.; Lega, M.A.; Feely, M.A.; Uphoff, T.S.; Jalal, S.M. Mosaic ring 20 with no detectable deletion by FISH analysis: Characteristic seizure disorder and literature review. Am. J. Med. Genet. 2006, 140, 1696–1706. [Google Scholar] [CrossRef] [PubMed]
- Ottaviani, A.; Schluth-Bolard, C.; Gilson, E.; Magdinier, F. D4Z4 as a prototype of CTCF and lamins-dependent insulator in human cells. Nucleus 2010, 1, 30–36. [Google Scholar] [CrossRef] [PubMed]
- Robin, J.D.; Ludlow, A.T.; Batten, K.; Gaillard, M.-C.; Stadler, G.; Magdinier, F.; Wright, W.E.; Shay, J.W. SORBS2 transcription is activated by telomere position effect–over long distance upon telomere shortening in muscle cells from patients with facioscapulohumeral dystrophy. Genome Res. 2015, 25, 1781–1790. [Google Scholar] [CrossRef] [PubMed]
- Gaillard, M.C.; Broucqsault, N.; Morere, J.; Laberthonniere, C.; Dion, C.; Badja, C.; Roche, S.; Nguyen, K.; Magdinier, F.; Robin, J. Analysis of the 4q35 chromatin organization reveals distinct long-range interactions in patients affected with Facio-Scapulo-Humeral Dystrophy. BioRxiv 2018. [Google Scholar] [CrossRef]
- Van den Boogaard, M.L.; Thijssen, P.E.; Aytekin, C.; Licciardi, F.; Kıykım, A.A.; Spossito, L.; Dalm, V.A.S.H.; Driessen, G.J.; Kersseboom, R.; de Vries, F.; et al. Expanding the mutation spectrum in ICF syndrome: Evidence for a gender bias in ICF2. Clin. Genet. 2017, 92, 380–387. [Google Scholar] [CrossRef] [PubMed]
- Xu, G.-L.; Bestor, T.H.; Bourc’his, D.; Hsieh, C.-L.; Tommerup, N.; Bugge, M.; Hulten, M.; Qu, X.; Russo, J.J.; Viegas-Péquignot, E. Chromosome instability and immunodeficiency syndrome caused by mutations in a DNA methyltransferase gene. Nature 1999, 402, 187–191. [Google Scholar] [CrossRef]
- De Greef, J.C.; Wang, J.; Balog, J.; den Dunnen, J.T.; Frants, R.R.; Straasheijm, K.R.; Aytekin, C.; van der Burg, M.; Duprez, L.; Ferster, A.; et al. Mutations in ZBTB24 are associated with immunodeficiency, centromeric instability, and facial anomalies syndrome type 2. Am. J. Hum. Genet. 2011, 88, 796–804. [Google Scholar] [CrossRef]
- Thijssen, P.E.; Ito, Y.; Grillo, G.; Wang, J.; Velasco, G.; Nitta, H.; Unoki, M.; Yoshihara, M.; Suyama, M.; Sun, Y.; et al. Mutations in CDCA7 and HELLS cause immunodeficiency–centromeric instability–facial anomalies syndrome. Nat. Commun. 2015, 6, 7870. [Google Scholar] [CrossRef] [Green Version]
- Deng, Z.; Campbell, A.E.; Lieberman, P.M. TERRA, CpG methylation, and telomere heterochromatin: Lessons from ICF syndrome cells. Cell Cycle 2010, 9, 69–74. [Google Scholar] [CrossRef] [Green Version]
- Yehezkel, S.; Segev, Y.; Viegas-Péquignot, E.; Skorecki, K.; Selig, S. Hypomethylation of subtelomeric regions in ICF syndrome is associated with abnormally short telomeres and enhanced transcription from telomeric regions. Hum. Mol. Genet. 2008, 17, 2776–2789. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Toubiana, S.; Velasco, G.; Chityat, A.; Kaindl, A.M.; Hershtig, N.; Tzur-Gilat, A.; Francastel, C.; Selig, S. Subtelomeric methylation distinguishes between subtypes of immunodeficiency, centromeric instability and facial anomalies syndrome. Hum. Mol. Genet. 2018, 27, 3568–3581. [Google Scholar] [CrossRef] [PubMed]
- De Sandre-Giovannoli, A. Lamin A truncation in hutchinson-gilford progeria. Science 2003, 300, 2055. [Google Scholar] [CrossRef] [PubMed]
- Taimen, P.; Pfleghaar, K.; Shimi, T.; Moller, D.; Ben-Harush, K.; Erdos, M.R.; Adam, S.A.; Herrmann, H.; Medalia, O.; Collins, F.S.; et al. A progeria mutation reveals functions for lamin A in nuclear assembly, architecture, and chromosome organization. Proc. Natl. Acad. Sci. USA 2009, 106, 20788–20793. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gonzalez-Suarez, I.; Redwood, A.B.; Perkins, S.M.; Vermolen, B.; Lichtensztejin, D.; Grotsky, D.A.; Morgado-Palacin, L.; Gapud, E.J.; Sleckman, B.P.; Sullivan, T.; et al. Novel roles for A-type lamins in telomere biology and the DNA damage response pathway. EMBO J. 2009, 28, 2414–2427. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Redwood, A.B.; Perkins, S.M.; Vanderwaal, R.P.; Feng, Z.; Biehl, K.J.; Gonzalez-Suarez, I.; Morgado-Palacin, L.; Shi, W.; Sage, J.; Roti-Roti, J.L.; et al. A dual role for A-type lamins in DNA double-strand break repair. Cell Cycle 2011, 10, 2549–2560. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- López-Otín, C.; Blasco, M.A.; Partridge, L.; Serrano, M.; Kroemer, G. The hallmarks of aging. Cell 2013, 153, 1194–1217. [Google Scholar] [CrossRef]
- Njajou, O.T.; Hsueh, W.-C.; Blackburn, E.H.; Newman, A.B.; Wu, S.-H.; Li, R.; Simonsick, E.M.; Harris, T.M.; Cummings, S.R.; Cawthon, R.M. Association between telomere length, specific causes of death, and years of healthy life in health, aging, and body composition, a population-based cohort study. J. Gerontol. 2009, 64, 860–864. [Google Scholar] [CrossRef]
- Aulinas, A.; Ramírez, M.-J.; Barahona, M.-J.; Valassi, E.; Resmini, E.; Mato, E.; Santos, A.; Crespo, I.; Bell, O.; Surrallés, J.; et al. Dyslipidemia and chronic inflammation markers are correlated with telomere length shortening in Cushing’s syndrome. PLoS ONE 2015, 10, e0120185. [Google Scholar] [CrossRef]
- Tellechea, M.L.; Pirola, C.J. The impact of hypertension on leukocyte telomere length: A systematic review and meta-analysis of human studies. J. Hum. Hypertens. 2017, 31, 99–105. [Google Scholar] [CrossRef]
- Willeit, P.; Willeit, J.; Brandstätter, A.; Ehrlenbach, S.; Mayr, A.; Gasperi, A.; Weger, S.; Oberhollenzer, F.; Reindl, M.; Kronenberg, F.; et al. Cellular aging reflected by leukocyte telomere length predicts advanced atherosclerosis and cardiovascular disease risk. Arterioscler. Thromb. Vasc. Biol. 2010, 30, 1649–1656. [Google Scholar] [CrossRef] [PubMed]
- Haycock, P.C.; Heydon, E.E.; Kaptoge, S.; Butterworth, A.S.; Thompson, A.; Willeit, P. Leucocyte telomere length and risk of cardiovascular disease: Systematic review and meta-analysis. BMJ 2014, 349. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Han, W.; Xue, W.; Zou, Y.; Xie, C.; Du, J.; Jin, G. The association between telomere length and cancer risk in population studies. Sci. Rep. 2016, 6, 22243. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, J.; Dong, X.; Cao, L.; Sun, Y.; Qiu, Y.; Zhang, Y.; Cao, R.; Covasa, M.; Zhong, L. Association between telomere length and diabetes mellitus: A meta-analysis. J. Int. Med. Res. 2016, 44, 1156–1173. [Google Scholar] [CrossRef] [PubMed] [Green Version]



© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Laberthonnière, C.; Magdinier, F.; Robin, J.D. Bring It to an End: Does Telomeres Size Matter? Cells 2019, 8, 30. https://doi.org/10.3390/cells8010030
Laberthonnière C, Magdinier F, Robin JD. Bring It to an End: Does Telomeres Size Matter? Cells. 2019; 8(1):30. https://doi.org/10.3390/cells8010030
Chicago/Turabian StyleLaberthonnière, Camille, Frédérique Magdinier, and Jérôme D. Robin. 2019. "Bring It to an End: Does Telomeres Size Matter?" Cells 8, no. 1: 30. https://doi.org/10.3390/cells8010030
APA StyleLaberthonnière, C., Magdinier, F., & Robin, J. D. (2019). Bring It to an End: Does Telomeres Size Matter? Cells, 8(1), 30. https://doi.org/10.3390/cells8010030