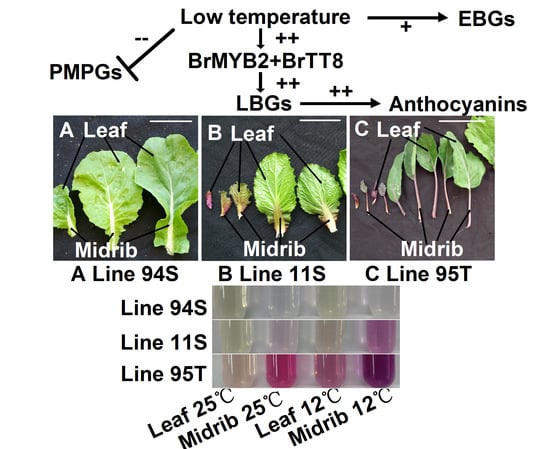
Low Temperature Promotes Anthocyanin Biosynthesis and Related Gene Expression in the Seedlings of Purple Head Chinese Cabbage (Brassica rapa L.)
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials and Growth Conditions
2.2. Histological Observation of Seedling Leaves
2.3. Total Anthocyanins Analysis
2.4. Identification of Anthocyanin Biosyntheis Genes and Quantitative Real-Time PCR (qRT-PCR) Analysis
2.5. Clustering Analysis and Interaction Network
2.6. Statistical Analysis
3. Results
3.1. Low Temperature Induces Anthocyanin Accumulation
3.2. Low Temperature Significantly Down-Regulated Most Phenylpropanoid Metabolic Pathway Genes (PMPGs)
3.3. Low Temperature Moderately Up-Regulated the EBGs for Subsequent Anthocyanin Biosynthesis
3.4. Key LBGs and Transport Genes Were Highly Induced under Low-Temperature Condition
3.5. Effect of Low Temperature on the Expression of Positive Regulatory Genes
3.6. Effect of Low Temperature on the Expression of Negative Regulatory Genes
3.7. Classification Scheme of Gene Expression Data and Interaction Networks
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Bi, H.T.; Guo, M.L.; Wang, J.W.; Qu, Y.; Du, W.L.; Zhang, K.M. Transcriptome analysis reveals anthocyanin acts as a protectant in Begonia semperflorens under low temperature. Acta Physiol. Plant. 2018, 40, 10–21. [Google Scholar] [CrossRef]
- Wiczkowski, W.; Topolska, J.; Honke, J. Anthocyanins profile and antioxidant capacity of red cabbages are influenced by genotype and vegetation period. J. Funct. Foods 2014, 7, 201–211. [Google Scholar] [CrossRef]
- Song, T.; Li, K.; Wu, T.; Wang, Y.; Zhang, X.; Xu, X.; Yao, Y.; Han, Z. Identification of new regulators through transcriptome analysis that regulate anthocyanin biosynthesis in apple leaves at low temperatures. PLoS ONE 2019, 14, e0210672. [Google Scholar] [CrossRef] [PubMed]
- Naing, A.H.; Kim, C.K. Roles of R2R3-MYB transcription factors in transcriptional regulation of anthocyanin biosynthesis in horticultural plants. Plant Mol. Biol. 2018, 98, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Macdonald, M.; D’Cunha, G. A modern view of phenylalanine ammonia lyase. Biochem. Cell Biol. 2007, 85, 273. [Google Scholar] [CrossRef]
- Li, Y.; Kim, J.I.; Pysh, L.; Chapple, C. Four isoforms of Arabidopsis thaliana 4-coumarate: CoA ligase (4CL) have overlapping yet distinct roles in phenylpropanoid metabolism. Plant Physiol. 2015, 169, 2409–2421. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.P.; Xu, Z.D.; Yu, X.Y.; Zhao, L.Y.; Zhao, M.Y.; Han, X.; Qi, S. Identification of two novel R2R3-MYB transcription factors, PsMYB114L and PsMYB12L, related to anthocyanin biosynthesis in Paeonia suffruticosa. Int. J. Mol. Sci. 2019, 20, 1055. [Google Scholar] [CrossRef] [Green Version]
- Sun, W.; Meng, X.; Liang, L.; Jiang, W.; Huang, Y.; He, J.; Hu, H.; Almqvist, J.; Gao, X.; Wang, L. Molecular and Biochemical Analysis of Chalcone Synthase from Freesia hybrid in Flavonoid Biosynthetic Pathway. PLoS ONE 2015, 10, e0119054. [Google Scholar] [CrossRef] [Green Version]
- Pelletier, M.K.; Shirley, B.W. Characterization of flavonol synthase and leucoanthocyanidin dioxygenase genes in Arabidopsis. Further evidence for differential regulation of “early” and “late” genes. Plant Physiol. 1997, 113, 1437. [Google Scholar] [CrossRef] [Green Version]
- Albert, N.W.; Davies, K.M.; Lewis, D.H.; Huaibi, Z.; Mirco, M.; Cyril, B.; Boase, M.R.; Hanh, N.; Jameson, P.E.; Schwinn, K.E. A conserved network of transcriptional activators and repressors regulates anthocyanin pigmentation in eudicots. Plant Cell 2014, 26, 962–980. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.J.; Hua, H.; Jiang, X.L.; Wang, P.Q.; Tao, X. A WD40 repeat protein from Camellia sinensis regulates anthocyanin and proanthocyanidin accumulation through the formation of MYB–bHLH–WD40 ternary complexes. Int. J. Mol. Sci. 2018, 19, 1686. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Petroni, K.; Tonelli, C. Recent advances on the regulation of anthocyanin synthesis in reproductive organs. Plant Sci. 2011, 181, 219–229. [Google Scholar] [CrossRef] [PubMed]
- Stracke, R.; Ishihara, H.; Huep, G.; Barsch, A.; Mehrtens, F.; Niehaus, K.; Weisshaar, B. Differential regulation of closely related R2R3-MYB transcription factors controls flavonol accumulation in different parts of the Arabidopsis thaliana seedling. Plant J. 2007, 50, 660–677. [Google Scholar] [CrossRef] [Green Version]
- Fu, Z.Z.; Wang, L.M.; Shang, H.Q.; Dong, X.Y.; Jiang, H.; Zhang, J.; Wang, H.J.; Li, Y.M.; Yuan, X.; Meng, S.Y. An R3-MYB gene of Phalaenopsis, MYBx1, represses anthocyanin accumulation. Plant Growth Regul. 2019, 88, 129–138. [Google Scholar] [CrossRef]
- Baudry, A.; Heim, M.A.; Dubreucq, B.; Caboche, M.; Weisshaar, B.; Lepiniec, L. TT2, TT8, and TTG1 synergistically specify the expression of BANYULS and proanthocyanidin biosynthesis in Arabidopsis thaliana. Plant J. 2004, 39, 366–380. [Google Scholar] [CrossRef]
- Dubos, C.; Le Gourrierec, J.; Baudry, A.; Huep, G.; Lanet, E.; Debeaujon, I.; Routaboul, J.M.; Alboresi, A.; Weisshaar, B.; Lepiniec, L. MYBL2 is a new regulator of flavonoid biosynthesis in Arabidopsis thaliana. Plant J. 2008, 55, 940–953. [Google Scholar] [CrossRef]
- Kyoko, M.; Yoshimi, U.; Masaru, O.T. AtMYBL2, a protein with a single MYB domain, acts as a negative regulator of anthocyanin biosynthesis in Arabidopsis. Plant J. 2010, 55, 954–967. [Google Scholar]
- Zhu, H.F.; Fitzsimmons, K.; Khandelwal, A.; Kranz, R.G. CPC, a single-repeat R3 MYB, is a negative regulator of anthocyanin biosynthesis in Arabidopsis. Mol. Plant 2009, 2, 790–802. [Google Scholar] [CrossRef]
- Rubin, G.; Tohge, T.; Matsuda, F.; Saito, K.; Scheible, W.R. Members of the LBD family of transcription factors repress anthocyanin synthesis and affect additional nitrogen responses in Arabidopsis. Plant Cell Online 2009, 21, 3567–3584. [Google Scholar] [CrossRef] [Green Version]
- Page, M.; Sultana, N.; Paszkiewicz, K.; Florance, H.; Smirnoff, N. The influence of ascorbate on anthocyanin accumulation during high light acclimation in Arabidopsis thaliana: Further evidence for redox control of anthocyanin synthesis. Plant Cell Environ. 2012, 35, 388–404. [Google Scholar] [CrossRef]
- Fang, H.C.; Dong, Y.H.; Yue, X.X.; Hu, J.F.; Jiang, S.H.; Xu, H.F.; Wang, Y.C.; Su, M.Y.; Zhang, J.; Zhang, Z.Y. The B-box zinc finger protein MdBBX20 integrates anthocyanin accumulation in response to ultraviolet radiation and low temperature. Plant Cell Environ. 2019, 42, 2090–2104. [Google Scholar] [CrossRef] [PubMed]
- Landi, M.; Pardossi, A.; Remorini, D.; Guidi, L. Antioxidant and photosynthetic response of a purple-leaved and a green-leaved cultivar of sweet basil (Ocimum basilicum) to boron excess. Environ. Exp. Bot. 2013, 85, 64–75. [Google Scholar] [CrossRef]
- Zhang, Y.T.; Liu, Y.; Hu, W.J.; Sun, B.; Chen, Q.; Tang, H.R. Anthocyanin accumulation and related gene expression affected by low temperature during strawberry coloration. Acta Physiol. Plant. 2018, 40, 192–199. [Google Scholar] [CrossRef]
- Zhang, C.; Jia, H.F.; Wu, W.M.; Wang, X.C.; Fang, J.G.; Wang, C. Functional conservation analysis and expression modes of grape anthocyanin synthesis genes responsive to low temperature stress. Gene 2015, 574, 168–177. [Google Scholar] [CrossRef]
- Gao-Takai, M.; Katayama-Ikegami, A.; Matsuda, K.; Shindo, H.; Uemae, S.; Oyaizu, M. A low temperature promotes anthocyanin biosynthesis but does not accelerate endogenous abscisic acid accumulation in red-skinned grapes. Plant Sci. 2019, 283, 165–176. [Google Scholar] [CrossRef]
- Tian, J.; Han, Z.Y.; Zhang, L.R.; Song, T.T.; Zhang, J.; Li, J.Y.; Yao, Y. Induction of anthocyanin accumulation in crabapple (Malus cv.) leaves by low temperatures. Hortsci. Publ. Am. Soc. Hortic. Sci. 2015, 50, 640–649. [Google Scholar] [CrossRef] [Green Version]
- Leng, P.; Qi, J.X. Effect of anthocyanin on David peach (Prunus davidiana Franch) under low temperature stress. Sci. Hortic. 2003, 97, 27–39. [Google Scholar] [CrossRef]
- Lo Piero, A.R.; Puglisi, I.; Rapisarda, P.; Petrone, G. Anthocyanins accumulation and related gene expression in red orange fruit induced by low temperature storage. J. Agric. Food Chem. 2005, 53, 9083–9088. [Google Scholar] [CrossRef]
- Wang, N.; Qu, C.; Jiang, S.; Chen, Z.; Xu, H.; Fang, H.; Su, M.; Zhang, J.; Wang, Y.; Liu, W.; et al. The proanthocyanidin-specific transcription factor MdMYBPA1 initiates anthocyanin synthesis under low-temperature conditions in red-fleshed apples. Plant J. 2018, 96, 39–55. [Google Scholar] [CrossRef] [Green Version]
- Ubi, B.E.; Honda, C.; Bessho, H.; Kondo, S.; Wada, M.; Kobayashi, S.; Moriguchi, T. Expression analysis of anthocyanin biosynthetic genes in apple skin: Effect of UV-B and temperature. Plant Sci. 2006, 170, 571–578. [Google Scholar] [CrossRef]
- Zhang, B.; Hu, Z.L.; Zhang, Y.J.; Li, Y.L.; Zhou, S.; Chen, G.P. A putative functional MYB transcription factor induced by low temperature regulates anthocyanin biosynthesis in purple kale (Brassica Oleracea var. acephala f. tricolor). Plant Cell Rep. 2012, 31, 281–289. [Google Scholar] [CrossRef] [PubMed]
- Landi, M.; Tattini, M.; Gould, K.S. Multiple functional roles of anthocyanins in plant-environment interactions. Environ. Exp. Bot. 2015, 119, 4–17. [Google Scholar] [CrossRef]
- Zhang, Y.Q.; Zheng, S.; Liu, Z.J.; Wang, L.G.; Bi, Y.R. Both HY5 and HYH are necessary regulators for low temperature-induced anthocyanin accumulation in Arabidopsis seedlings. J. Plant Physiol. 2011, 168, 367–374. [Google Scholar] [CrossRef] [PubMed]
- Sun, B.M.; Zhu, Z.S.; Cao, P.R.; Chen, H.; Chen, C.M.; Zhou, X.; Mao, Y.H.; Lei, J.J.; Jiang, Y.P.; Meng, W.; et al. Purple foliage coloration in tea (Camellia sinensis L.) arises from activation of the R2R3-MYB transcription factor CsAN1. Sci. Rep. 2016, 6, 32534. [Google Scholar] [CrossRef]
- Guo, N.; Cheng, F.; Wu, J.; Liu, B.; Zheng, S.N.; Liang, J.L.; Wang, X.W. Anthocyanin biosynthetic genes in Brassica rapa. BMC Genom. 2014, 15, 426. [Google Scholar] [CrossRef] [Green Version]
- Ahmed, N.U.; Park, J.I.; Jung, H.J.; Hur, Y.; Nou, I.S. Anthocyanin biosynthesis for cold and freezing stress tolerance and desirable color in Brassica rapa. Funct. Integr. Genom. 2015, 15, 383–394. [Google Scholar] [CrossRef]
- Ahmed, N.U.; Park, J.I.; Jung, H.J.; Yang, T.J.; Hur, Y.; Nou, I.S. Characterization of dihydroflavonol 4-reductase (DFR) genes and their association with cold and freezing stress in Brassica rapa. Gene 2014, 550, 46–55. [Google Scholar] [CrossRef]
- Lux, A.; Morita, S.; Abe, J.; Ito, K. An improved method for clearing and staining free-hand sections and whole-mount samples. Ann. Bot. 2005, 96, 989–996. [Google Scholar] [CrossRef]
- He, Q.; Zhang, Z.F.; Zhang, L.G. Anthocyanin Accumulation, Antioxidant Ability and Stability, and a Transcriptional Analysis of Anthocyanin Biosynthesis in Purple Heading Chinese Cabbage (Brassica rapa L. ssp. pekinensis). J. Agric. Food Chem. 2016, 64, 132–145. [Google Scholar] [CrossRef]
- Giusti, M.M.; Wrolstad, R.E. Characterization and Measurement of Anthocyanins by UV-Visible Spectroscopy. In Current Protocols in Food Analytical Chemistry; Unit F1.2; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2001; pp. 1–13. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2-△△CT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Caraux, G.; Pinloche, S. PermutMatrix: A graphical environment to arrange gene expression profiles in optimal linear order. Bioinformatics 2005, 21, 1280–1281. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Szklarczyk, D.; Morris, J.H.; Cook, H.; Kuhn, M.; Wyder, S.; Simonovic, M.; Santos, A.; Doncheva, N.T.; Roth, A.; Bork, P. The STRING database in 2017: Quality-controlled protein–protein association networks, made broadly accessible. Nucleic Acids Res. 2017, 45, D362–D368. [Google Scholar] [CrossRef] [PubMed]
- Gou, J.Y.; Felippes, F.F.; Liu, C.J.; Weigel, D.; Wang, J.W. Negative regulation of anthocyanin biosynthesis in Arabidopsis by a miR156-targeted SPL transcription factor. Plant Cell Online 2011, 23, 1512–1522. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fraser, C.M.; Thompson, M.G.; Shirley, A.M.; Ralph, J.; Schoenherr, J.A.; Sinlapadech, T.; Hall, M.C.; Chapple, C. Related Arabidopsis serine carboxypeptidase-like sinapoylglucose acyltransferases display distinct but overlapping substrate specificities. Plant Physiol. 2007, 144, 1986. [Google Scholar] [CrossRef] [Green Version]
- Keiko, Y.S.; Fukushima, A.; Nakabayashi, R.; Hanada, K.; Matsuda, F.; Sugawara, S.; Inoue, E.; Kuromori, T.; Ito, T.; Shinozaki, K. Two glycosyltransferases involved in anthocyanin modification delineated by transcriptome independent component analysis in Arabidopsis thaliana. Plant J. 2012, 69, 154–167. [Google Scholar]
- Meißner, D.; Albert, A.; Böttcher, C.; Strack, D.; Milkowski, C. The role of UDP-glucose:hydroxycinnamate glucosyltransferases in phenylpropanoid metabolism and the response to UV-B radiation in Arabidopsis thaliana. Planta 2008, 228, 663. [Google Scholar] [CrossRef]
- Carmona, L.; Alquezar, B.; Marques, V.V.; Pena, L. Anthocyanin biosynthesis and accumulation in blood oranges during postharvest storage at different low temperatures. Food Chem. 2017, 237, 7–14. [Google Scholar] [CrossRef]
- Ban, Y.; Honda, C.; Hatsuyama, Y.; Igarashi, M.; Bessho, H.; Moriguchi, T. Isolation and functional analysis of a MYB transcription factor gene that is a key regulator for the development of red coloration in apple skin. Plant Cell Physiol. 2007, 48, 958–970. [Google Scholar] [CrossRef]
- Xie, X.B.; Li, S.; Zhang, R.F.; Zhao, J.; Chen, Y.C.; Zhao, Q.; Yao, Y.X.; You, C.X.; Zhang, X.S.; Hao, Y.J. The bHLH transcription factor MdbHLH3 promotes anthocyanin accumulation and fruit colouration in response to low temperature in apples. Plant Cell Environ. 2012, 35, 1884–1897. [Google Scholar] [CrossRef]
- Crifo, T.; Petrone, G.; Lo Cicero, L.; Lo Piero, A.R. Short cold storage enhances the anthocyanin contents and level of transcripts related to their biosynthesis in blood oranges. J. Agric. Food Chem. 2012, 60, 476–481. [Google Scholar] [CrossRef]
- Zhou, L.J.; Li, Y.Y.; Zhang, R.F.; Zhang, C.L.; Xie, X.B.; Zhao, C.; Hao, Y.J. The small ubiquitin-like modifier E3 ligase MdSIZ1 promotes anthocyanin accumulation by sumoylating MdMYB1 under low-temperature conditions in apple. Plant Cell Environ. 2017, 40, 2068–2080. [Google Scholar] [CrossRef] [PubMed]
- Hajime, H.; Tomoko, F.A.; Toshikatsu, O.; Minoru, N.; Masahiko, S. Anthocyanin accumulation and related gene expression in Japanese parsley (Oenanthe stolonifera, DC.) induced by low temperature. J. Plant Physiol. 2001, 158, 71–78. [Google Scholar]
- Rowan, D.D.; Cao, M.; Kui, L.W.; Cooney, J.M.; Jensen, D.J.; Austin, P.T.; Hunt, M.B.; Norling, C.; Hellens, R.P.; Schaffer, R.J. Environmental regulation of leaf colour in red 35S:PAP1 Arabidopsis thaliana. New Phytol. 2009, 182, 102–115. [Google Scholar] [CrossRef]
- Tian, J.; Chen, M.; Zhang, J.; Li, K.; Song, T.; Zhang, X.; Yao, Y. Characteristics of dihydroflavonol 4-reductase gene promoters from different leaf colored Malus crabapple cultivars. Hortic. Res. 2017, 4, 17070. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sakai, M.; Yamagishi, M.; Matsuyama, K. Repression of anthocyanin biosynthesis by R3-MYB transcription factors in lily (Lilium spp.). Plant Cell Rep. 2019, 38, 609–622. [Google Scholar] [CrossRef] [PubMed]
- Albert, N.W.; Lewis, D.H.; Zhang, H.; Irving, L.J.; Jameson, P.E.; Davies, K.M. Light-induced vegetative anthocyanin pigmentation in Petunia. J. Exp. Bot. 2009, 60, 2191–2202. [Google Scholar] [CrossRef] [Green Version]
- Spelt, C.; Quattrocchio, F.; Mol, J.N.; Koes, R. Anthocyanin1 of petunia encodes a basic helix-loop-helix protein that directly activates transcription of structural anthocyanin genes. Plant Cell Online 2000, 12, 1619–1631. [Google Scholar] [CrossRef] [Green Version]
- Albert, N.W.; Lewis, D.H.; Huaibi, Z.; Schwinn, K.E.; Jameson, P.E.; Davies, K.M. Members of an R2R3-MYB transcription factor family in Petunia are developmentally and environmentally regulated to control complex floral and vegetative pigmentation patterning. Plant J. 2011, 65, 771–784. [Google Scholar] [CrossRef]
- Shang, Y.J.; Julien, V.; Steve, M.; Bailey, P.C.; Schwinn, K.E.; Jameson, P.E.; Martin, C.R.; Davies, K.M. The molecular basis for venation patterning of pigmentation and its effect on pollinator attraction in flowers of Antirrhinum. New Phytol. 2011, 189, 602–615. [Google Scholar] [CrossRef]
- Hiroyoshi, K.; Mutsumi, K.; Koji, G. Expression analysis of ANTHOCYANINLESS2 gene in Arabidopsis. Plant Sci. 2009, 175, 853–857. [Google Scholar]
- Song, H.; Yi, H.; Lee, M.; Han, C.T.; Lee, J.; Kim, H.R.; Park, J.I.; Nou, I.S.; Kim, S.J.; Hur, Y. Purple Brassica oleracea var. capitata F. rubra is due to the loss of BoMYBL2–1 expression. BMC Plant Biol. 2018, 18, 82. [Google Scholar] [PubMed]
- Wang, Y.C.; Sun, J.J.; Wang, N.; Xu, H.F.; Chen, X.S. MdMYBL2 helps regulate cytokinin-induced anthocyanin biosynthesis in red-fleshed apple (Malus sieversii f. niedzwetzkyana) callus. Funct. Plant Biol. 2019, 46, 187–196. [Google Scholar] [CrossRef]
- Li, H.H.; Liu, X.; An, J.P.; Hao, Y.J.; Wang, X.F.; You, C.X. Cloning and elucidation of the functional role of apple MdLBD13 in anthocyanin biosynthesis and nitrate assimilation. Plant Cell Tissue Organ Cult. 2017, 130, 1–13. [Google Scholar] [CrossRef]









© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
He, Q.; Ren, Y.; Zhao, W.; Li, R.; Zhang, L. Low Temperature Promotes Anthocyanin Biosynthesis and Related Gene Expression in the Seedlings of Purple Head Chinese Cabbage (Brassica rapa L.). Genes 2020, 11, 81. https://doi.org/10.3390/genes11010081
He Q, Ren Y, Zhao W, Li R, Zhang L. Low Temperature Promotes Anthocyanin Biosynthesis and Related Gene Expression in the Seedlings of Purple Head Chinese Cabbage (Brassica rapa L.). Genes. 2020; 11(1):81. https://doi.org/10.3390/genes11010081
Chicago/Turabian StyleHe, Qiong, Yanjing Ren, Wenbin Zhao, Ru Li, and Lugang Zhang. 2020. "Low Temperature Promotes Anthocyanin Biosynthesis and Related Gene Expression in the Seedlings of Purple Head Chinese Cabbage (Brassica rapa L.)" Genes 11, no. 1: 81. https://doi.org/10.3390/genes11010081
APA StyleHe, Q., Ren, Y., Zhao, W., Li, R., & Zhang, L. (2020). Low Temperature Promotes Anthocyanin Biosynthesis and Related Gene Expression in the Seedlings of Purple Head Chinese Cabbage (Brassica rapa L.). Genes, 11(1), 81. https://doi.org/10.3390/genes11010081