Novel Aspects on The Interaction Between Grapevine and Plasmopara viticola: Dual-RNA-Seq Analysis Highlights Gene Expression Dynamics in The Pathogen and The Plant During The Battle For Infection
Abstract
:1. Introduction
2. Materials and Methods
2.1. RNA-seq Data
2.2. P. viticola Transcriptome: Read Preparation, Alignment, and Analysis of Differentially Expressed Genes
2.3. GO Enrichments for P. viticola DEGs
2.4. Network Analysis of Grapevine Transcriptome: Gene Coexpression Network Analysis
2.5. Network Analysis of Grapevine Transcriptome: Functional Enrichment Analyses
2.6. Network Analysis of Grapevine Transcriptome: Transcription Factor Enrichment
2.7. De novo Transcriptome Assembly of Grapevine Cultivars
2.8. Statistical Analysis of Differentially Expressed De novo Transcripts
2.9. Availability of Data and Materials
3. Results
3.1. P. viticola Transcriptome Analysis
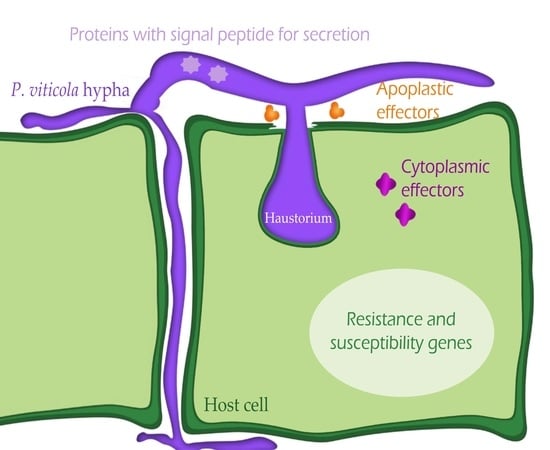
3.2. P. viticola Effectors
3.3. Grapevine Transcriptome Network Analysis
3.4. De novo Grapevine Transcriptome Assembly
3.5. Unique Gene Expression Patterns of Grapevine Cultivars Under Infection
4. Discussion
4.1. P. viticola Transcriptome
4.2. P. viticola Effectors
4.3. Networking Analysis Reveals A Putative Gene of Susceptibility in V. vinifera
4.4. De novo Transcriptome Assembly Reveals New Grapevine Traits Related to P. viticola Response
4.5. Transcriptomic Changes Occurring In Grapevine During The First Stages of P. viticola Infection
4.6. Unique DEGs of Mgaloblishvili in Response to P. viticola Infection
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Jacobsen, B.J. Role of plant pathology in Integrated Pest Management. Annu. Rev. Phytopathol. 1997, 35, 373–391. [Google Scholar] [CrossRef]
- Molitor, D.; Biewers, B.; Junglen, M.; Schultz, M.; Clement, P.; Permesang, G.; Regnery, D.; Porten, M.; Herzog, K.; Hoffmann, L.; et al. Multi-annual comparisons demonstrate differences in the bunch rot susceptibility of nine Vitis vinifera L. ’Riesling’ clones. Vitis 2018, 57, 17–25. [Google Scholar]
- Rotolo, C.; De Miccolis Angelini, R.M.; Dongiovanni, C.; Pollastro, S.; Fumarola, G.; Di Carolo, M.; Perrelli, D.; Natale, P.; Faretra, F. Use of biocontrol agents and botanicals in integrated management of Botrytis cinerea in table grape vineyards. Pest Manag. Sci. 2018, 74, 715–725. [Google Scholar] [CrossRef]
- Passera, A.; Compant, S.; Casati, P.; Maturo, M.G.; Battelli, G.; Quaglino, F.; Antonielli, L.; Salerno, D.; Brasca, M.; Toffolatti, S.L.; et al. Not just a pathogen? Description of a plant-beneficial Pseudomonas syringae strain. Front. Microbiol. 2019, 10, 1409. [Google Scholar] [CrossRef] [Green Version]
- Toffolatti, S.L.; Russo, G.; Campia, P.; Bianco, P.A.; Borsa, P.; Coatti, M.; Torriani, S.F.; Sierotzki, H. A time-course investigation of resistance to the carboxylic acid amide mandipropamid in field populations of Plasmopara viticola treated with anti-resistance strategies. Pest Manag. Sci. 2018, 74, 2822–2834. [Google Scholar] [CrossRef]
- Boyd, L.A.; Ridout, C.; O’Sullivan, D.M.; Leach, J.E.; Leung, H. Plant–pathogen interactions: Disease resistance in modern agriculture. Trends Genet. 2013, 29, 233–240. [Google Scholar] [CrossRef]
- REX Consortium. Combining selective pressures to enhance the durability of disease resistance genes. Front. Plant Sci. 2016, 7, 1916. [Google Scholar]
- Zaidi, S.S.; Mukhtar, M.S.; Mansoor, S. Genome editing: Targeting susceptibility genes for plant disease resistance. Trends Biotechnol. 2018, 36, 898–906. [Google Scholar] [CrossRef] [PubMed]
- McGovern, P.E.; Jalabadze, M.; Batiuk, S.; Callahan, M.P.; Smith, K.E.; Hall, G.R.; Kvavadze, E.; Maghradze, D.; Rusishvili, N.; Bouby, L.; et al. Early Neolithic wine of Georgia in the South Caucasus. Proc. Natl. Acad. Sci. USA 2017, 114, E10309–E10318. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Emmett, R.W.; Wicks, T.J.; Magarey, P.A. Downy mildew of grapes. In Plant Diseases of International Importance—Diseases of Fruit Crops; Kumar, J., Chaube, H.S., Singh, U.S., Mukhopadhyay, A.N., Eds.; Prentice All: Englewood Cliffs, NJ, USA, 1992; Volume III, pp. 90–128. [Google Scholar]
- Gessler, C.; Pertot, I.; Perazzolli, M. Plasmopara viticola: A review of knowledge on downy mildew of grapevine and effective disease management. Phytopathol. Mediterr. 2011, 50, 3–44. [Google Scholar]
- Buonassisi, D.; Colombo, M.; Migliaro, D.; Dolzani, C.; Peressotti, E.; Mizzotti, C.; Velasco, R.; Masiero, S.; Perazzolli, M.; Vezzulli, S. Breeding for grapevine downy mildew resistance: A review of “omics” approaches. Euphytica 2017, 213, 103. [Google Scholar] [CrossRef]
- Merdinoglu, D.; Schneider, C.; Prado, E.; Wiedemann-Merdinoglu, S.; Mestre, P. Breeding for durable resistance to downy and powdery mildew in grapevine. OENO One 2018, 52, 203–209. [Google Scholar] [CrossRef] [Green Version]
- Boubals, D. Contribution a l’etude des causes de la resistance des Vitacees au mildiou de la vigne (Plasmopara viticola (B. et C.) Berl. et de T.) et leur mode de transmission hereditaire. Ann. Amélior. Plant. 1959, 9, 5–233. [Google Scholar]
- Pedneault, K.; Provost, C. Fungus resistant grape varieties as a suitable alternative for organic wine production: Benefits, limits, and challenges. Sci. Hortic. 2016, 208, 57–77. [Google Scholar] [CrossRef]
- Toffolatti, S.L.; Maddalena, G.; Salomoni, D.; Maghradze, D.; Bianco, P.A.; Failla, O. Evidence of resistance to the downy mildew agent Plasmopara viticola in the Georgian Vitis vinifera germplasm. Vitis 2016, 55, 121–128. [Google Scholar]
- Toffolatti, S.L.; De Lorenzis, G.; Costa, A.; Maddalena, G.; Passera, A.; Bonza, M.C.; Pindo, M.; Stefani, E.; Cestaro, A.; Casati, P.; et al. Unique resistance traits against downy mildew from the center of origin of grapevine (Vitis vinifera). Sci. Rep. 2018, 8, 12523. [Google Scholar] [CrossRef]
- Rovenich, H.; Boshoven, J.C.; Thomma, B.P.H.J. Filamentous pathogen effector functions: Of pathogens, hosts and microbiomes. Curr. Opin. Plant Biol. 2014, 20, 96–103. [Google Scholar] [CrossRef] [Green Version]
- Kamoun, S. Groovy times: Filamentous pathogen effectors revealed. Curr. Opin. Plant Biol. 2007, 10, 358–365. [Google Scholar] [CrossRef]
- Franceschetti, M.; Maqbool, A.; Jiménez-Dalmaroni, M.J.; Pennington, H.G.; Kamoun, S.; Banfield, M.J. Effectors of filamentous plant pathogens: Commonalities amid diversity. Microbiol. Mol. Biol. Rev. 2017, 81, e00066-16. [Google Scholar] [CrossRef] [Green Version]
- Brilli, M.; Asquini, E.; Moser, M.; Bianchedi, P.L.; Perazzolli, M.; Si-Ammour, A. A multi-omics study of the grapevine-downy mildew (Plasmopara viticola) pathosystem unveils a complex protein coding and noncoding-based arms race during infection. Sci. Rep. 2018, 8, 757. [Google Scholar] [CrossRef] [Green Version]
- Mestre, P.; Piron, M.C.; Merdinoglou, D. Identification of effector genes from the phytopathogenic Oomycete Plasmopara viticola through the analysis of gene expression in germinated zoospores. Fungal Biol. 2012, 116, 825–835. [Google Scholar] [CrossRef] [Green Version]
- Mestre, P.; Carrere, S.; Gouzy, J.; Piron, M.-C.; Tourvieille de Labrouhee, D.; Vincourt, P.; Delmotte, F.; Godiard, L. Comparative analysis of expressed CRN and RXLR effectors from two Plasmopara species causing grapevine and sunflower downy mildew. Plant Pathol. 2016, 65, 767–781. [Google Scholar] [CrossRef]
- Xiang, J.; Li, X.; Wu, J.; Yin, L.; Zhang, Y.; Lu, J. Studying the mechanism of Plasmopara viticola RxLR effectors on suppressing plant immunity. Front. Microbiol. 2016, 7, 709. [Google Scholar] [CrossRef]
- Xiang, J.; Li, X.; Yin, L.; Liu, Y.; Zhang, Y.; Qu, J.; Lu, J. A candidate RxLR effector from Plasmopara viticola can elicit immune responses in Nicotiana benthamiana. BMC Plant Biol. 2017, 17, 75. [Google Scholar] [CrossRef] [Green Version]
- Yin, L.; Li, X.; Xiang, J.; Qu, J.; Zhang, Y.; Dry, I.B.; Lu, J. Characterization of the secretome of Plasmopara viticola by de novo transcriptome analysis. Physiol. Mol. Plant Pathol. 2015, 91, 1–10. [Google Scholar] [CrossRef]
- Yin, L.; An, Y.; Qu, J.; Li, X.; Zhang, Y.; Dry, I.; Wu, H.; Lu, J. Genome sequence of Plasmopara viticola and insight into the pathogenic mechanism. Sci. Rep. 2017, 7, 46553. [Google Scholar] [CrossRef] [Green Version]
- Stam, R.; Jupe, J.; Howden, A.J.M.; Morris, J.A.; Boevink, P.C.; Hedley, P.E.; Huitema, E. Identification and characterisation of CRN effectors in Phytophthora capsici shows modularity and functional diversity. PLoS ONE 2013, 8, e59517. [Google Scholar] [CrossRef]
- Amaro, T.M.M.M.; Thilliez, G.J.A.; Motion, G.B.; Huitema, E. A perspective on CRN proteins in the genomics age: Evolution, classification, delivery and function revisited. Front. Plant Sci. 2017, 8, 99. [Google Scholar] [CrossRef] [Green Version]
- Lévesque, C.A.; Brouwer, H.; Cano, L.; Hamilton, J.P.; Holt, C.; Huitema, E.; Raffaele, S.; Robideau, G.P.; Thines, M.; Win, J.; et al. Genome sequence of the necrotrophic plant pathogen Pythium ultimum reveals original pathogenicity mechanisms and effector repertoire. Genome Biol. 2010, 11, R73. [Google Scholar] [CrossRef]
- McGowan, J.; Fitzpatrick, D.A. Genomic, network, and phylogenetic analysis of the oomycete effector arsenal. mSphere 2017, 2, e00408-17. [Google Scholar] [CrossRef] [Green Version]
- Leesutthiphonchai, W.; Vu, A.L.; Ah-Fong, A.M.V.; Judelson, H.S. How does Phytophthora infestans evade control efforts? Modern insight into the late blight disease. Phytopathology 2018, 108, 916–924. [Google Scholar] [CrossRef] [Green Version]
- Petre, B.; Kamoun, S. How do filamentous pathogens deliver effector proteins into plant cells? PLoS Biol. 2014, 12, e1001801. [Google Scholar] [CrossRef] [Green Version]
- Wang, S.; Boevink, P.C.; Welsh, L.; Zhang, R.; Whisson, S.C.; Birch, P.R.J. Delivery of cytoplasmic and apoplastic effectors from Phytophthora infestans haustoria by distinct secretion pathways. New Phytol. 2017, 216, 205–215. [Google Scholar] [CrossRef] [Green Version]
- Kamoun, S. A catalogue of the effector secretome of plant pathogenic Oomycetes. Annu. Rev. Phytopathol. 2006, 44, 41–60. [Google Scholar] [CrossRef] [Green Version]
- Kanneganti, T.-D.; Huitema, E.; Cakir, C.; Kamoun, S. Synergistic interactions of the plant cell death pathways induced by Phytophthora infestans Nep1-Like Protein PiNPP1.1 and INF1 Elicitin. MPMI 2006, 19, 854–863. [Google Scholar] [CrossRef] [Green Version]
- Dangl, J.L.; Horvath, D.M.; Staskawicz, B.J. Pivoting the plant immune system from dissection to deployment. Science 2013, 341, 746–751. [Google Scholar] [CrossRef] [Green Version]
- Giraldo, M.C.; Valent, B. Filamentous plant pathogen effectors in action. Nat. Rev. Microbiol. 2013, 11, 800–814. [Google Scholar] [CrossRef]
- Miller, R.N.G.; Alves, G.S.C.; Van Sluys, M.-A. Plant immunity: Unravelling the complexity of plant responses to biotic stresses. Ann. Bot. 2017, 119, 681–687. [Google Scholar] [CrossRef] [Green Version]
- Plasmopara viticola Multi Omics Dissection. Available online: https://www.researchgate.net/project/Plasmopara-viticola-multi-omics-dissection (accessed on 1 February 2018).
- Tian, T.; Liu, Y.; Yan, H.; You, Q.; Yi, X.; Du, Z.; Xu, W.; Su, Z. agriGO v2.0: A GO analysis toolkit for the agricultural community, 2017 update. Nucleic Acids Res. 2017, 3, 45. [Google Scholar] [CrossRef]
- Supek, F.; Bošnjak, M.; Škunca, N.; Šmuc, T. REVIGO summarizes and visualizes long lists of Gene Ontology terms. PLoS ONE 2011, 6, e21800. [Google Scholar] [CrossRef] [Green Version]
- Bardou, P.; Mariette, J.; Escudié, F.; Djemiel, C.; Klopp, C. jvenn: An interactive Venn diagram viewer. BMC Bioinform. 2014, 15, 293. [Google Scholar] [CrossRef] [Green Version]
- Warnes, G.R.; Bolker, B.; Lumley, T. gplots: Various R Programming Tools for Plotting Data. 2009. Available online: http://cran.r-project.org/web/packages/gplots/index.html (accessed on 25 February 2020).
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-Seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [Green Version]
- Langfelder, P.; Horvath, S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinform. 2008, 9, 559. [Google Scholar] [CrossRef] [Green Version]
- Ravasz, E.; Somera, A.L.; Mongru, D.A.; Oltvai, Z.N.; Barabasi, A.L. Hierarchical organization of modularity in metabolic networks. Science 2002, 297, 1551–1555. [Google Scholar] [CrossRef] [Green Version]
- Langfelder, P. Signed vs. Unsigned Topological Overlap Matrix. Technical Report. 2013. Available online: https://horvath.genetics.ucla.edu/html/CoexpressionNetwork/Rpackages/WGCNA/TechnicalReports/signedTOM.pdf (accessed on 7 November 2018).
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef] [Green Version]
- The Kore cluster. Available online: http://sit.fbk.eu/hpc/kore (accessed on 1 April 2009).
- Fu, L.; Niu, B.; Zhu, Z.; Wu, S.; Li, W. CD-HIT: Accelerated for clustering the next-generation sequencing data. Bioinformatics 2012, 28, 3150–3152. [Google Scholar] [CrossRef]
- Conesa, A.; Götz, S.; García-Gómez, J.M.; Terol, J.; Talón, M.; Robles, M. Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 2005, 21, 3674–3676. [Google Scholar] [CrossRef] [Green Version]
- Langmead, B.; Salzberg, S. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [Green Version]
- Di Gaspero, G.; Cipriani, G. Resistance gene analogs are candidate markers for disease-resistance genes in grape (Vitis spp.). Theor. Appl. Genet. 2002, 106, 163–172. [Google Scholar] [CrossRef]
- Blackman, L.M.; Cullerne, D.P.; Hardham, A.R. Bioinformatic characterisation of genes encoding cell wall degrading enzymes in the Phytophthora parasitica genome. BMC Genom. 2014, 15, 785. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Toffolatti, S.L.; Venturini, G.; Maffi, D.; Vercesi, A. Phenotypic and histochemical traits of the interaction between Plasmopara viticola and resistant or susceptible grapevine varieties. BMC Plant Biol. 2012, 12, 124. [Google Scholar] [CrossRef] [Green Version]
- Gauthier, A.; Trouvelot, S.; Kelloniemi, J.; Frettinger, P.; Wendehenne, D.; Daire, X.; Joubert, J.M.; Ferrarini, A.; Delledonne, M.; Flors, V.; et al. The sulfated laminarin triggers a stress transcriptome before priming the SA- and ROS-dependent defenses during grapevine’s induced resistance against Plasmopara viticola. PLoS ONE 2014, 9, e88145. [Google Scholar] [CrossRef]
- Legay, G.; Marouf, E.; Berger, D.; Neuhaus, J.-M.; Mauch-Mani, B.; Slaughter, A. Identification of genes expressed during the compatible interaction of grapevine with Plasmopara viticola through suppression subtractive hybridization (SSH). Eur. J. Plant Pathol. 2001, 129, 281–301. [Google Scholar] [CrossRef] [Green Version]
- Martin, K.; McDougall, B.M.; McIlroy, S.; Jayus Chen, J.; Seviour, R.J. Biochemistry and molecular biology of exocellular fungal β-(1,3)- and β-(1,6)-glucanases. FEMS Microbiol. Rev. 2007, 31, 168–192. [Google Scholar] [CrossRef]
- Valdés-Santiago, L.; Cervantes-Chávez, J.A.; León-Ramírez, C.G.; Ruiz-Herrera, J. Polyamine metabolism in fungi with emphasis on phytopathogenic species. J. Amino Acids 2012, 2012, 837932. [Google Scholar] [CrossRef] [Green Version]
- Majumdar, R.; Lebar, M.; Mackm, B.; Minocha, R.; Minocha, S.; Carter-Wientjes, C.; Sickler, C.; Rajasekaran, K.; Cary, J.W. The Aspergillus flavus Spermidine Synthase (spds) Gene, is required for normal development, aflatoxin production, and pathogenesis during infection of maize kernels. Front. Plant Sci. 2018, 9, 317. [Google Scholar] [CrossRef] [Green Version]
- Sakihama, Y.; Mano, J.; Sano, S.; Asada, K.; Yamasaki, H. Reduction of Phenoxyl Radicals mediated by Monodehydroascorbate Reductase. Biochem. Biophys. Res. Commun. 2000, 279, 949–954. [Google Scholar] [CrossRef]
- Wojtaszek, P. Oxidative burst: An early plant response to pathogen infection. Biochem. J. 1997, 322, 681–692. [Google Scholar] [CrossRef] [Green Version]
- Ah-Fong, A.M.V.; Su Kim, K.; Judelson, H.S. RNA-seq of life stages of the oomycete Phytophthora infestans reveals dynamic changes in metabolic, signal transduction, and pathogenesis genes and a major role for calcium signaling in development. BMC Genom. 2017, 18, 198. [Google Scholar] [CrossRef] [Green Version]
- Aliaga, G.R.; Ellzey, J.T. Ultrastructural localization of acid phosphatase and alkaline phosphatase within oogonia of Achlya recurve. Mycologia 1984, 76, 85–98. [Google Scholar] [CrossRef]
- Lapin, D.; Van den Ackerveken, G. Susceptibility to plant disease: More than a failure of host immunity. Trends Plant Sci. 2014, 18, 546–554. [Google Scholar] [CrossRef]
- Chitarrini, G.; Soini, E.; Riccadonna, S.; Franceschi, P.; Zulini, L.; Masuero, D.; Vecchione, A.; Stefanini, M.; Di Gaspero, G.; Mattivi, F.; et al. Identification of biomarkers for defense response to Plasmopara viticola in a resistant grape variety. Front. Plant Sci. 2017, 8, 1524. [Google Scholar] [CrossRef]
- Fawke, S.; Doumane, M.; Schornack, S. Oomycete interactions with plants: Infection strategies and resistance principles. Microbiol. Mol. Biol. Rev. 2015, 79, 263–279. [Google Scholar] [CrossRef] [Green Version]
- Pavan, S.; Jacobsen, E.; Visser, R.G.F.; Bai, Y. Loss of susceptibility as a novel breeding strategy for durable and broad-spectrum resistance. Mol. Breed. 2010, 25, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Van Schie, C.C.N.; Takken, F.L.W. Susceptibility Genes 101: How to Be a Good Host. Annu. Rev. Phytopathol. 2014, 52, 551–581. [Google Scholar] [CrossRef]
- Weis, C.; Pfeilmeier, S.; Glawischnig, E.; Isono, E.; Pachl, F.; Hahne, H.; Kuster, B.; Eichmann, R.; Hückelhoven, R. Co-immunoprecipitation-based identification of putative BAX INHIBITOR-1-interacting proteins involved in cell death regulation and plant-powdery mildew interactions. Mol. Plant Pathol. 2013, 14, 791–802. [Google Scholar] [CrossRef]
- Thatcher, L.F.; Powell, J.J.; Aitken, E.A.; Kazan, K.; Manners, J.M. The lateral organ boundaries domain transcription factor LBD20 functions in Fusarium wilt susceptibility and jasmonate signaling in Arabidopsis. Plant Physiol. 2012, 160, 407–418. [Google Scholar] [CrossRef] [Green Version]
- This, P.; Lacombe, T.; Thomas, M.R. Historical origins and genetic diversity of wine grapes. Trends Genet. 2006, 22, 511–519. [Google Scholar] [CrossRef]
- Corso, M.; Vannozzi, A.; Maza, E.; Vitulo, N.; Meggio, F.; Pitacco, A.; Telatin, A.; D’Angelo, M.; Feltrin, E.; Negri, A.S.; et al. Comprehensive transcript profiling of two grapevine rootstock genotypes contrasting in drought susceptibility links the phenylpropanoid pathway to enhanced tolerance. J. Exp. Bot. 2015, 66, 5739–5752. [Google Scholar] [CrossRef]
- Vannozzi, A.; Donnini, S.; Vigani, G.; Corso, M.; Valle, G.; Vitulo, N.; Bonghi, C.; Zocchi, G.; Lucchin, M. Transcriptional characterization of a widely-used grapevine rootstock genotype under different iron-limited conditions. Front. Plant Sci. 2016, 7, 1994. [Google Scholar] [CrossRef] [Green Version]
- Weng, K.; Li, Z.-Q.; Liu, R.-Q.; Wang, L.; Wang, Y.-J.; Xu, Y. Transcriptome of Erysiphe necator-infected Vitis pseudoreticulata leaves provides insight into grapevine resistance to powdery mildew. Hortic. Res. 2014, 1, 14049. [Google Scholar] [CrossRef] [Green Version]
- Yang, Y.; Mao, L.; Jittayasothorn, Y.; Kang, Y.; Jiao, C.; Fei, Z.; Zhong, G.-Y. Messenger RNA exchange between scions and rootstocks in grafted grapevines. BMC Plant Biol. 2015, 15, 251. [Google Scholar] [CrossRef] [Green Version]
- Vitulo, N.; Forcato, C.; Carpinelli, E.C.; Telatin, A.; Campagna, D.; D’Angelo, M.; Zimbello, R.; Corso, M.; Vannozzi, A.; Bonghi, C.; et al. A deep survey of alternative splicing in grape reveals changes in the splicing machinery related to tissue, stress condition and genotype. BMC Plant Biol. 2014, 14, 99. [Google Scholar] [CrossRef] [Green Version]
- Ardito, F.; Giuliani, M.; Perrone, D.; Troiano, G.; Lo Muzio, L. The crucial role of protein phosphorylation in cell signaling and its use as targeted therapy (Review). Int. J. Mol. Med. 2017, 40, 271–280. [Google Scholar] [CrossRef] [Green Version]
- Xing, T.; Ouellet, T.; Miki, B.L. Towards genomic and proteomic studies of protein phosphorylation in plant-pathogen interactions. Trends Plant Sci. 2002, 7, 224–230. [Google Scholar] [CrossRef]
- Yang, Y.; Shah, J.; Klessig, D.F. Signal perception and transduction in plant defense responses. Genes Dev. 1997, 11, 1621–1639. [Google Scholar] [CrossRef] [Green Version]
- Yangnan, G.; Zavaliev, R.; Dong, X. Membrane trafficking in plant immunity. Mol. Plant. 2017, 10, 1026–1034. [Google Scholar]
- Dangl, J.L.; Jones, J.D.G. Plant pathogens and integrated defence responses to infection. Nature 2001, 411, 826–833. [Google Scholar] [CrossRef]
- Bellin, D.; Peressotti, E.; Merdinoglu, D.; Wiedemann-Merdinoglu, S.; Adam-Blondon, A.-F.; Cipriani, G.; Morgante, M.; Testolin, R.; Di Gaspero, G. Resistance to Plasmopara viticola in grapevine “Bianca” is controlled by a major dominant gene causing localised necrosis at the infection site. Theor. Appl. Genet. 2009, 120, 163–176. [Google Scholar] [CrossRef] [Green Version]
- Di Gaspero, G.; Copetti, D.; Coleman, C.; Castellarin, S.D.; Eibach, R.; Kozma, P.; Lacombe, T.; Gambetta, G.; Zvyagin, A.; Cindrić, P.; et al. Selective sweep at the Rpv3 locus during grapevine breeding for downy mildew resistance. Theor. Appl. Genet. 2012, 124, 277–286. [Google Scholar] [CrossRef]
- Fröbel, S.; Dudenhöffer, J.; Töpfer, R.; Zyprian, E. Transcriptome analysis of early downy mildew (Plasmopara viticola) defense in grapevines carrying the Asian resistance locus Rpv10. Euphytica 2019, 215, 28. [Google Scholar] [CrossRef]
- Polesani, M.; Bortesi, L.; Ferrarini, A.; Zamboni, A.; Fasoli, M.; Zadra, C.; Lovato, A.; Pezzotti, M.; Delledonne, M.; Polverari, A. General and species-specific transcriptional responses to downy mildew infection in a susceptible (Vitis vinifera) and a resistant (V. riparia) grapevine species. BMC Genom. 2010, 11, 117. [Google Scholar] [CrossRef] [Green Version]
- Jürges, G.; Kassemeyer, H.H.; Durrenberger, M.; Duggelin, M.; Nick, P. The mode of interaction between Vitis and Plasmopara viticola Berk. & Curt. Ex de Bary depends on the host species. Plant Biol. 2009, 11, 886–898. [Google Scholar]
- Peressotti, E.; Wiedemann-Merdinoglu, S.; Delmotte, F.; Bellin, D.; Di Gaspero, G.; Testolin, R.; Merdinoglu, D.; Mestre, P. Breakdown of resistance to grapevine downy mildew upon limited deployment of a resistant variety. BMC Plant Biol. 2010, 10, 147. [Google Scholar] [CrossRef] [Green Version]
- Afzal, A.J.; Wood, A.J.; Lightfoot, D.A. Plant receptor-like serine threonine kinases: Roles in signaling and plant defense. Mol. Plant Microbe Interact. 2008, 21, 507–517. [Google Scholar] [CrossRef] [Green Version]
- Whitham, S.A.; Yang, C.; Goodin, M.M. Global impact: Elucidating plant responses to viral infection. Mol. Plant Microbe Interact. 2006, 19, 1207–1215. [Google Scholar] [CrossRef] [Green Version]
- Maimbo, M.; Ohnishi, K.; Hikichi, Y.; Yoshioka, H.; Kiba, A. Induction of a small heat shock protein and its functional roles in Nicotiana plants in the defense response against Ralstonia solanacearum. Plant Physiol. 2007, 145, 1588–1599. [Google Scholar] [CrossRef] [Green Version]
- Fang, X.; Chen, W.; Xin, Y.; Zhang, H.; Yan, C.; Yu, H.; Liu, H.; Xiao, W.; Wang, S.; Zheng, G.; et al. Proteomic analysis of strawberry leaves infected with Colletotrichum fragariae. J. Proteom. 2012, 75, 4074–4090. [Google Scholar] [CrossRef]
- Manter, D.K.; Karchesy, J.J.; Kelsey, R.G. The sporicidal activity of yellow-cedar heartwood, essential oil and wood constituents toward Phytophthora ramorum in culture. Forest Pathol. 2006, 36, 297–308. [Google Scholar] [CrossRef]
- Sharon-Asa, L.; Shalit, M.; Frydman, A.; Bar, E.; Holland, D.; Or, E.; Lavi, U.; Lewinsohn, E.; Eyal, Y. Citrus fruit flavor and aroma biosynthesis: Isolation, functional characterization, and developmental regulation of Cstps1, a key gene in the production of the sesquiterpene aroma compound valencene. Plant J. 2003, 36, 664–674. [Google Scholar] [CrossRef]
- Lücker, J.; Bowen, P.; Bohlmann, J. Vitis vinifera terpenoid cyclases: Functional identification of two sesquiterpene synthase cDNAs encoding (+)-valencene synthase and (−)-germacrene D synthase and expression of mono- and sesquiterpene synthases in grapevine flowers and berries. Phytochemistry 2004, 65, 2649–2659. [Google Scholar] [CrossRef]
- Nishitani, K.; Vissenberg, K. Roles of the XTH protein family in the expanding cell. In The Expanding Cell - Plant Cell Monographs; Verbelen, J.-P., Vissenberg, K., Eds.; Springer: Berlin, Germany, 2007; Volume 5, pp. 89–116. [Google Scholar]
- Maris, A.; Suslov, D.; Fry, S.C.; Verbelen, J.-P.; Vissenberg, K. Enzymic characterization of two recombinant xyloglucan endotransglucosylase/hydrolase (XTH) proteins of Arabidopsis and their effect on root growth and cell wall extension. J. Exp. Bot. 2009, 60, 3959–3972. [Google Scholar] [CrossRef] [Green Version]
- Han, Y.; Ban, Q.; Li, H.; Hou, Y.; Jin, M.; Han, S.; Rao, J. DkXTH8, a novel xyloglucan endotransglucosylase/hydrolase in persimmon, alters cell wall structure and promotes leaf senescence and fruit postharvest softening. Sci. Rep. 2016, 6, 39155. [Google Scholar] [CrossRef]
- Laureano, G.; Figueiredo, J.; Cavaco, A.R.; Duarte, B.; Caçador, I.; Malhó, R.; Sousa Silva, M.; Matos, A.R.; Figueiredo, A. The interplay between membrane lipids and phospholipase A family members in grapevine resistance against Plasmopara viticola. Sci. Rep. 2018, 8, 14538. [Google Scholar] [CrossRef]
- Pessina, S.; Lenzi, L.; Perazzolli, M.; Campa, M.; Dalla Costa, L.; Urso, S.; Valè, G.; Salamini, F.; Velasco, R.; Malnoy, M. Knockdown of MLO genes reduces susceptibility to powdery mildew in grapevine. Hortic. Res. 2016, 20, 16016. [Google Scholar] [CrossRef] [Green Version]




| DEGs | Contrast | Total | ||||||
|---|---|---|---|---|---|---|---|---|
| PvM_vs_PvP | PvM_vs_PvB | PvP_vs_PvB | ||||||
| Nr. | % | Nr. | % | Nr. | % | Nr. | % | |
| Up | 1 | 0.065 | 524 | 34.5 | 939 | 61.82 | 1464 | 96.39 |
| Down | 1 | 0.065 | 9 | 0.59 | 45 | 2.96 | 55 | 3.62 |
| Total | 2 | 0.13 | 533 | 35.09 | 984 | 64.78 | 1519 | 100 |
| Library Run accession (Study PRJEB24540; European Nucleotide Archive) | Library Type (Cultivar Name, Treatment, dai Number) | Number of Reads Passing Trimmomatic Filter for Trinity de novo Assembly | Number of Transcripts Assembled by Trinity | Number of Supertranscripts Assembled by Trinity |
|---|---|---|---|---|
| ERR2274751, ERR2274752, ERR2274753 | Mgaloblishvili, non-inoculated, 0 dai | 77,329,414 | 48,263 | 33,701 |
| ERR2274754, ERR2274755, ERR2274756 | Mgaloblishvili, non-inoculated, 1 dai | 77,053,567 | 43,102 | 31,513 |
| ERR2274757, ERR2274758, ERR2274759 | Mgaloblishvili, non-inoculated, 2 dai | 79,505,005 | 47,912 | 33,969 |
| ERR2274760, ERR2274761, ERR2274762 | Mgaloblishvili, non-inoculated, 3 dai | 59,610,375 | 44,152 | 32,075 |
| ERR2276774, ERR2276775, ERR2276776 | Mgaloblishvili, inoculated, 1 dai | 80,263,242 | 50,930 | 34,701 |
| ERR2276777, ERR2276778, ERR2276779 | Mgaloblishvili, inoculated, 2 dai | 74,298,111 | 51,109 | 35,146 |
| ERR2276780, ERR2276781, ERR2276782 | Mgaloblishvili, inoculated, 3 dai | 86,910,171 | 43,151 | 31,905 |
| ERR2262368, ERR2262369, ERR2262534 | Pinot noir, non-inoculated, 0 dai | 77,508,661 | 38,749 | 28,818 |
| ERR2265572, ERR2265573, ERR2265574 | Pinot noir, non-inoculated, 1 dai | 71,622,301 | 44,433 | 32,403 |
| ERR2265575, ERR2265576, ERR2265577 | Pinot noir, non-inoculated, 2 dai | 71,910,056 | 45,555 | 32,779 |
| ERR2265578, ERR2265579, ERR2265580 | Pinot noir, non-inoculated, 3 dai | 72,205,904 | 47,860 | 35,603 |
| ERR2262591, ERR2262592, ERR2262593 | Pinot noir, inoculated, 1 dai | 72,980,600 | 40,313 | 30,198 |
| ERR2264780, ERR2264781, ERR2264782 | Pinot noir, inoculated, 2 dai | 72,188,628 | 47,313 | 34,683 |
| ERR2265538, ERR2265539, ERR2265540 | Pinot noir, inoculated, 3 dai | 81,804,077 | 45,398 | 34,396 |
| ERR2271536, ERR2271537, ERR2271538 | Bianca, non-inoculated, 0 dai | 83,267,610 | 55,833 | 35,120 |
| ERR2271539, ERR2271540, ERR2271541 | Bianca, non-inoculated, 1 dai | 73,434,003 | 40,946 | 30,225 |
| ERR2271542, ERR2271543, ERR2271544 | Bianca, non-inoculated, 2 dai | 81,386,614 | 54,370 | 34,995 |
| ERR2271545, ERR2271546, ERR2271547 | Bianca, non-inoculated, 3 dai | 79,917,013 | 59,852 | 37,236 |
| ERR2276787, ERR2276788, ERR2276789 | Bianca, inoculated, 1 dai | 78,491,558 | 57,665 | 36,349 |
| ERR2276790, ERR2276791, ERR2276792 | Bianca, inoculated, 2 dai | 79,960,138 | 56,124 | 35,554 |
| ERR2276793, ERR2276794, ERR2276795 | Bianca, inoculated, 3 dai | 84,125,204 | 60,459 | 37,891 |
| Cultivar | Differentially Expressed Genes | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 dai | 2 dai | 3 dai | |||||||
| Nr. | % | Nr. | % | Nr. | % | ||||
| Mgaloblishvili | |||||||||
| Up | 2592 | 4.20 | 162 | 0.26 | 73 | 0.12 | |||
| Down | 3976 | 6.50 | 40 | 0.06 | 87 | 0.14 | |||
| Pinot noir | |||||||||
| Up | 1349 | 2.50 | 769 | 1.30 | 4905 | 8.50 | |||
| Down | 1349 | 2.50 | 145 | 0.25 | 4551 | 7.80 | |||
| Bianca | |||||||||
| Up | 6564 | 8.90 | 256 | 0.34 | 5789 | 7.50 | |||
| Down | 9884 | 13.0 | 228 | 0.30 | 5327 | 6.90 | |||
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Toffolatti, S.L.; De Lorenzis, G.; Brilli, M.; Moser, M.; Shariati, V.; Tavakol, E.; Maddalena, G.; Passera, A.; Casati, P.; Pindo, M.; et al. Novel Aspects on The Interaction Between Grapevine and Plasmopara viticola: Dual-RNA-Seq Analysis Highlights Gene Expression Dynamics in The Pathogen and The Plant During The Battle For Infection. Genes 2020, 11, 261. https://doi.org/10.3390/genes11030261
Toffolatti SL, De Lorenzis G, Brilli M, Moser M, Shariati V, Tavakol E, Maddalena G, Passera A, Casati P, Pindo M, et al. Novel Aspects on The Interaction Between Grapevine and Plasmopara viticola: Dual-RNA-Seq Analysis Highlights Gene Expression Dynamics in The Pathogen and The Plant During The Battle For Infection. Genes. 2020; 11(3):261. https://doi.org/10.3390/genes11030261
Chicago/Turabian StyleToffolatti, Silvia Laura, Gabriella De Lorenzis, Matteo Brilli, Mirko Moser, Vahid Shariati, Elahe Tavakol, Giuliana Maddalena, Alessandro Passera, Paola Casati, Massimo Pindo, and et al. 2020. "Novel Aspects on The Interaction Between Grapevine and Plasmopara viticola: Dual-RNA-Seq Analysis Highlights Gene Expression Dynamics in The Pathogen and The Plant During The Battle For Infection" Genes 11, no. 3: 261. https://doi.org/10.3390/genes11030261
APA StyleToffolatti, S. L., De Lorenzis, G., Brilli, M., Moser, M., Shariati, V., Tavakol, E., Maddalena, G., Passera, A., Casati, P., Pindo, M., Cestaro, A., Maghradze, D., Failla, O., Bianco, P. A., & Quaglino, F. (2020). Novel Aspects on The Interaction Between Grapevine and Plasmopara viticola: Dual-RNA-Seq Analysis Highlights Gene Expression Dynamics in The Pathogen and The Plant During The Battle For Infection. Genes, 11(3), 261. https://doi.org/10.3390/genes11030261