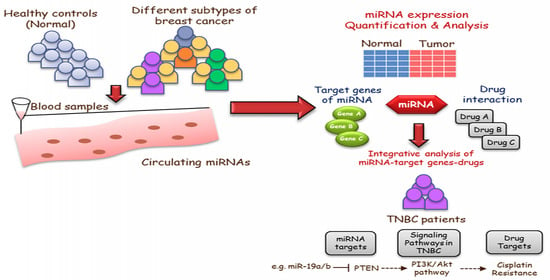
Clinical Identification of Dysregulated Circulating microRNAs and Their Implication in Drug Response in Triple Negative Breast Cancer (TNBC) by Target Gene Network and Meta-Analysis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Patient Specimen Accrual, Sample Collection, and Study Design
2.2. Isolation of Total RNA and miRNAs Analysis
2.3. Data Analysis and Bioinformatics
2.4. Gene-Set Enrichment (GSEA) and Drug Prediction Analysis
3. Results
3.1. Patient Characteristics
3.2. Circulating microRNA Expression Profile in Human TNBC Samples
3.3. Analysis of the Function of the Targets of Significant Circulating miRNAs in Triple-Negative Breast Cancer
3.4. Significant TNBC-Specific miRNAs Regulating Genes Involved in Cancer Drug Resistance
3.5. Association of Significant Circulating miRNAs with the Survival of Breast Cancer Patients
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Najminejad, H.; Kalantar, S.M.; Abdollahpour-Alitappeh, M.; Karimi, M.H.; Seifalian, A.M.; Gholipourmalekabadi, M.; Sheikhha, M.H. Emerging Roles of Exosomal MiRNAs in Breast Cancer Drug Resistance. IUBMB Life 2019, 71, 1672–1684. [Google Scholar] [CrossRef] [PubMed]
- Si, W.; Shen, J.; Zheng, H.; Fan, W. The Role and Mechanisms of Action of MicroRNAs in Cancer Drug Resistance. Clin. Epigenetics 2019, 11, 25. [Google Scholar] [CrossRef] [PubMed]
- Dass, S.A.; Tan, K.L.; Selva Rajan, R.; Mokhtar, N.F.; Adzmi, E.R.; Wan Abdul Rahman, W.F.; Tengku Din, T.A.D.A.-A.; Balakrishnan, V. Triple Negative Breast Cancer: A Review of Present and Future Diagnostic Modalities. Medicina 2021, 57, 62. [Google Scholar] [CrossRef]
- Baffa, R.; Fassan, M.; Volinia, S.; O’Hara, B.; Liu, C.-G.; Palazzo, J.P.; Gardiman, M.; Rugge, M.; Gomella, L.G.; Croce, C.M.; et al. MicroRNA Expression Profiling of Human Metastatic Cancers Identifies Cancer Gene Targets. J. Pathol. 2009, 219, 214–221. [Google Scholar] [CrossRef]
- Cho, W.C.S. OncomiRs: The Discovery and Progress of MicroRNAs in Cancers. Mol. Cancer 2007, 6, 60. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zelli, V.; Compagnoni, C.; Capelli, R.; Cannita, K.; Sidoni, T.; Ficorella, C.; Capalbo, C.; Zazzeroni, F.; Tessitore, A.; Alesse, E. Circulating MicroRNAs as Prognostic and Therapeutic Biomarkers in Breast Cancer Molecular Subtypes. J. Pers. Med. 2020, 10, 98. [Google Scholar] [CrossRef] [PubMed]
- Köberle, V.; Pleli, T.; Schmithals, C.; Augusto Alonso, E.; Haupenthal, J.; Bönig, H.; Peveling-Oberhag, J.; Biondi, R.M.; Zeuzem, S.; Kronenberger, B.; et al. Differential Stability of Cell-Free Circulating MicroRNAs: Implications for Their Utilization as Biomarkers. PLoS ONE 2013, 8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dean-Colomb, W.; Esteva, F.J. Her2-Positive Breast Cancer: Herceptin and Beyond. Eur. J. Cancer 2008, 44, 2806–2812. [Google Scholar] [CrossRef]
- Dunnwald, L.K.; Rossing, M.A.; Li, C.I. Hormone Receptor Status, Tumor Characteristics, and Prognosis: A Prospective Cohort of Breast Cancer Patients. Breast Cancer Res. 2007, 9, R6. [Google Scholar] [CrossRef]
- Dawson, S.J.; Provenzano, E.; Caldas, C. Triple Negative Breast Cancers: Clinical and Prognostic Implications. Eur. J. Cancer 2009, 45, 27–40. [Google Scholar] [CrossRef]
- Mehanna, J.; Haddad, F.G.; Eid, R.; Lambertini, M.; Kourie, H.R. Triple-Negative Breast Cancer: Current Perspective on the Evolving Therapeutic Landscape. Int. J. Women’s Heal. 2019, 11, 431–437. [Google Scholar] [CrossRef] [Green Version]
- Nedeljković, M.; Damjanović, A. Mechanisms of Chemotherapy Resistance in Triple-Negative Breast Cancer—How We Can Rise to the Challenge. Cells 2019, 8, 957. [Google Scholar] [CrossRef] [Green Version]
- Wu, N.; Zhang, J.; Zhao, J.; Mu, K.; Zhang, J.; Jin, Z.; Yu, J.; Liu, J. Precision Medicine Based on Tumorigenic Signaling Pathways for Triple-Negative Breast Cancer. Oncol. Lett. 2018, 16, 4984–4996. [Google Scholar] [CrossRef] [Green Version]
- Jayaraj, R.; Nayagam, S.G.; Kar, A.; Sathyakumar, S.; Mohammed, H.; Smiti, M.; Sabarimurugan, S.; Kumarasamy, C.; Priyadharshini, T.; Gothandam, K.M.; et al. Clinical Theragnostic Relationship between Drug-Resistance Specific MiRNA Expressions, Chemotherapeutic Resistance, and Sensitivity in Breast Cancer: A Systematic Review and Meta-Analysis. Cells 2019, 8, 1250. [Google Scholar] [CrossRef] [Green Version]
- Holubekova, V.; Kolkova, Z.; Grendar, M.; Brany, D.; Dvorska, D.; Stastny, I.; Jagelkova, M.; Zelinova, K.; Samec, M.; Liskova, A.; et al. Pathway Analysis of Selected Circulating MiRNAs in Plasma of Breast Cancer Patients: A Preliminary Study. Int. J. Mol. Sci. 2020, 21, 7288. [Google Scholar] [CrossRef]
- Kleivi Sahlberg, K.; Bottai, G.; Naume, B.; Burwinkel, B.; Calin, G.A.; Borresen-Dale, A.-L.; Santarpia, L. A Serum MicroRNA Signature Predicts Tumor Relapse and Survival in Triple-Negative Breast Cancer Patients. Clin. Cancer Res. 2015, 21, 1207–1214. [Google Scholar] [CrossRef] [Green Version]
- Kahraman, M.; Röske, A.; Laufer, T.; Fehlmann, T.; Backes, C.; Kern, F.; Kohlhaas, J.; Schrörs, H.; Saiz, A.; Zabler, C.; et al. MicroRNA in Diagnosis and Therapy Monitoring of Early-Stage Triple-Negative Breast Cancer. Sci. Rep. 2018, 8, 11584. [Google Scholar] [CrossRef]
- Hanna, J.; Hossain, G.S.; Kocerha, J. The Potential for MicroRNA Therapeutics and Clinical Research. Front. Genet. 2019, 10. [Google Scholar] [CrossRef] [Green Version]
- Yu, A.-M.; Jian, C.; Yu, A.H.; Tu, M.-J. RNA Therapy: Are We Using the Right Molecules? Pharmacol. Ther. 2019, 196, 91–104. [Google Scholar] [CrossRef]
- Golan, T.; Khvalevsky, E.Z.; Hubert, A.; Gabai, R.M.; Hen, N.; Segal, A.; Domb, A.; Harari, G.; David, E.B.; Raskin, S.; et al. RNAi Therapy Targeting KRAS in Combination with Chemotherapy for Locally Advanced Pancreatic Cancer Patients. Oncotarget 2015, 6, 24560–24570. [Google Scholar] [CrossRef] [Green Version]
- Qattan, A.; Intabli, H.; Alkhayal, W.; Eltabache, C.; Tweigieri, T.; Amer, S.B. Robust Expression of Tumor Suppressor MiRNA’s Let-7 and MiR-195 Detected in Plasma of Saudi Female Breast Cancer Patients. BMC Cancer 2017, 17, 799. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Gu, Z.; Eils, R.; Schlesner, M. Complex Heatmaps Reveal Patterns and Correlations in Multidimensional Genomic Data. Bioinformatics 2016, 32, 2847–2849. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blighe, K.; Rana, S.; Turkes, E.; Ostendorf, B.; Lewis, M. EnhancedVolcano: Publication-Ready Volcano Plots with Enhanced Colouring and Labeling. 2020. R Package Version 1.8.0. Available online: https://github.com/kevinblighe/EnhancedVolcano (accessed on 17 February 2021).
- Wickham, H.; Averick, M.; Bryan, J.; Chang, W.; McGowan, L.D.; François, R.; Grolemund, G.; Hayes, A.; Henry, L.; Hester, J.; et al. Welcome to the Tidyverse. J. Open Source Softw. 2019, 4, 1686. [Google Scholar] [CrossRef]
- Gu, Z.; Gu, L.; Eils, R.; Schlesner, M.; Brors, B. Circlize Implements and Enhances Circular Visualization in R. Bioinformatics 2014, 30, 2811–2812. [Google Scholar] [CrossRef] [Green Version]
- Wickham, H. Ggplot2: Elegant Graphics for Data AnalysisIn Use R! Springer: New York, NY, USA, 2009; ISBN 978-0-387-98141-3. [Google Scholar]
- Grossman, R.L.; Heath, A.P.; Ferretti, V.; Varmus, H.E.; Lowy, D.R.; Kibbe, W.A.; Staudt, L.M. Toward a Shared Vision for Cancer Genomic Data. N. Engl. J. Med. 2016, 375, 1109–1112. [Google Scholar] [CrossRef]
- Wishart, D.S.; Knox, C.; Guo, A.C.; Shrivastava, S.; Hassanali, M.; Stothard, P.; Chang, Z.; Woolsey, J. DrugBank: A Comprehensive Resource for in Silico Drug Discovery and Exploration. Nucleic Acids Res. 2006, 34, D668–D672. [Google Scholar] [CrossRef]
- Wishart, D.S.; Knox, C.; Guo, A.C.; Cheng, D.; Shrivastava, S.; Tzur, D.; Gautam, B.; Hassanali, M. DrugBank: A Knowledgebase for Drugs, Drug Actions and Drug Targets. Nucleic Acids Res. 2008, 36, D901–D906. [Google Scholar] [CrossRef]
- Knox, C.; Law, V.; Jewison, T.; Liu, P.; Ly, S.; Frolkis, A.; Pon, A.; Banco, K.; Mak, C.; Neveu, V.; et al. DrugBank 3.0: A Comprehensive Resource for “omics” Research on Drugs. Nucleic Acids Res. 2011, 39, D1035–D1041. [Google Scholar] [CrossRef] [Green Version]
- Barghout, S.H.; Zepeda, N.; Vincent, K.; Azad, A.K.; Xu, Z.; Yang, C.; Steed, H.; Postovit, L.-M.; Fu, Y. RUNX3 Contributes to Carboplatin Resistance in Epithelial Ovarian Cancer Cells. Gynecol. Oncol. 2015, 138, 647–655. [Google Scholar] [CrossRef]
- Gupta, I.; Rizeq, B.; Vranic, S.; Moustafa, A.-E.A.; Al Farsi, H. Circulating MiRNAs in HER2-Positive and Triple Negative Breast Cancers: Potential Biomarkers and Therapeutic Targets. Int. J. Mol. Sci. 2020, 21, 6750. [Google Scholar] [CrossRef]
- Anfossi, S.; Giordano, A.; Gao, H.; Cohen, E.N.; Tin, S.; Wu, Q.; Garza, R.J.; Debeb, B.G.; Alvarez, R.H.; Valero, V.; et al. High Serum MiR-19a Levels Are Associated with Inflammatory Breast Cancer and Are Predictive of Favorable Clinical Outcome in Patients with Metastatic HER2+ Inflammatory Breast Cancer. PLoS ONE 2014, 9, e83113. [Google Scholar] [CrossRef]
- Chakraborty, C.; Sharma, A.R.; Sharma, G.; Lee, S.-S. The Interplay among MiRNAs, Major Cytokines, and Cancer-Related Inflammation. Mol. Ther. Nucleic Acids 2020, 20, 606–620. [Google Scholar] [CrossRef]
- Mathe, A.; Scott, R.J.; Avery-Kiejda, K.A. MiRNAs and Other Epigenetic Changes as Biomarkers in Triple Negative Breast Cancer. Int J. Mol. Sci 2015, 16, 28347–28376. [Google Scholar] [CrossRef] [Green Version]
- Toyama, T.; Kondo, N.; Endo, Y.; Sugiura, H.; Yoshimoto, N.; Iwasa, M.; Takahashi, S.; Fujii, Y.; Yamashita, H. High Expression of MicroRNA-210 Is an Independent Factor Indicating a Poor Prognosis in Japanese Triple-Negative Breast Cancer Patients. Jpn J. Clin. Oncol 2012, 42, 256–263. [Google Scholar] [CrossRef] [Green Version]
- Souza, K.C.B.; Evangelista, A.F.; Leal, L.F.; Souza, C.P.; Vieira, R.A.; Causin, R.L.; Neuber, A.C.; Pessoa, D.P.; Passos, G.A.S.; Reis, R.M.V.; et al. Identification of Cell-Free Circulating MicroRNAs for the Detection of Early Breast Cancer and Molecular Subtyping. J. Oncol. 2019, 2019, 8393769. [Google Scholar] [CrossRef] [Green Version]
- Chen, H.; Pan, H.; Qian, Y.; Zhou, W.; Liu, X. MiR-25-3p Promotes the Proliferation of Triple Negative Breast Cancer by Targeting BTG2. Mol. Cancer 2018, 17. [Google Scholar] [CrossRef]
- Chan, J.J.; Tan, T.J.Y.; Dent, R.A. Novel Therapeutic Avenues in Triple-Negative Breast Cancer: PI3K/AKT Inhibition, Androgen Receptor Blockade, and Beyond. Adv. Med. Oncol. 2019, 11. [Google Scholar] [CrossRef]
- Lee, J.S.; Yost, S.E.; Yuan, Y. Neoadjuvant Treatment for Triple Negative Breast Cancer: Recent Progresses and Challenges. Cancers 2020, 12. [Google Scholar] [CrossRef]
- Yang, L.; Cai, Y.; Zhang, D.; Sun, J.; Xu, C.; Zhao, W.; Jiang, W.; Pan, C. MiR-195/MiR-497 Regulate CD274 Expression of Immune Regulatory Ligands in Triple-Negative Breast Cancer. J. Breast Cancer 2018, 21, 371. [Google Scholar] [CrossRef]
- Li, N.; Miao, Y.; Shan, Y.; Liu, B.; Li, Y.; Zhao, L.; Jia, L. MiR-106b and MiR-93 Regulate Cell Progression by Suppression of PTEN via PI3K/Akt Pathway in Breast Cancer. Cell Death Dis. 2017, 8, e2796. [Google Scholar] [CrossRef]
- Tang, Q.; Ouyang, H.; He, D.; Yu, C.; Tang, G. MicroRNA-Based Potential Diagnostic, Prognostic and Therapeutic Applications in Triple-Negative Breast Cancer. Artif. Cells Nanomed. Biotechnol. 2019, 47, 2800–2809. [Google Scholar] [CrossRef]
- Hu, J.; Xu, J.; Wu, Y.; Chen, Q.; Zheng, W.; Lu, X.; Zhou, C.; Jiao, D. Identification of MicroRNA-93 as a Functional Dysregulated MiRNA in Triple-Negative Breast Cancer. Tumour Biol. 2015, 36, 251–258. [Google Scholar] [CrossRef]
- Li, H.-Y.; Liang, J.-L.; Kuo, Y.-L.; Lee, H.-H.; Calkins, M.J.; Chang, H.-T.; Lin, F.-C.; Chen, Y.-C.; Hsu, T.-I.; Hsiao, M.; et al. MiR-105/93-3p Promotes Chemoresistance and Circulating MiR-105/93-3p Acts as a Diagnostic Biomarker for Triple Negative Breast Cancer. Breast Cancer Res. 2017, 19, 133. [Google Scholar] [CrossRef]
- Cai, Z.; Zhang, F.; Chen, W.; Zhang, J.; Li, H. MiRNAs: A Promising Target in the Chemoresistance of Bladder Cancer. OncoTargets Ther. 2020, 12, 11805–11816. [Google Scholar] [CrossRef] [Green Version]
- Fornari, F.; Milazzo, M.; Chieco, P.; Negrini, M.; Calin, G.A.; Grazi, G.L.; Pollutri, D.; Croce, C.M.; Bolondi, L.; Gramantieri, L.; et al. MiR-199a-3p Regulates MTOR and c-Met to Influence the Doxorubicin Sensitivity of Human Hepatocarcinoma Cells. Cancer Res. 2010, 70, 5184–5193. [Google Scholar] [CrossRef] [Green Version]
- Rodriguez, R.; Meuth, M. Chk1 and P21 Cooperate to Prevent Apoptosis during DNA Replication Fork Stress. Mol. Biol. Cell 2006, 17, 402–412. [Google Scholar] [CrossRef] [Green Version]
- Gartel, A.L.; Radhakrishnan, S.K. Lost in Transcription: P21 Repression, Mechanisms, and Consequences. Cancer Res. 2005, 65, 3980–3985. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barr, A.R.; Cooper, S.; Heldt, F.S.; Butera, F.; Stoy, H.; Mansfeld, J.; Novák, B.; Bakal, C. DNA Damage during S-Phase Mediates the Proliferation-Quiescence Decision in the Subsequent G1 via P21 Expression. Nat. Commun. 2017, 8, 14728. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Xu, Z. MiR-93 Enhances Cell Proliferation by Directly Targeting CDKN1A in Nasopharyngeal Carcinoma. Oncol. Lett. 2018, 15, 1723–1727. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, K.-L.; Kuo, Y.-C.; Ho, Y.-S.; Huang, Y.-H. Triple-Negative Breast Cancer: Current Understanding and Future Therapeutic Breakthrough Targeting Cancer Stemness. Cancers 2019, 11, 1334. [Google Scholar] [CrossRef] [Green Version]
- De Angelis, M.L.; Francescangeli, F.; Zeuner, A. Breast Cancer Stem Cells as Drivers of Tumor Chemoresistance, Dormancy and Relapse: New Challenges and Therapeutic Opportunities. Cancers 2019, 11, 1569. [Google Scholar] [CrossRef] [Green Version]
- Song, S.J.; Poliseno, L.; Song, M.S.; Ala, U.; Webster, K.; Ng, C.; Beringer, G.; Brikbak, N.J.; Yuan, X.; Cantley, L.C.; et al. MicroRNA-Antagonism Regulates Breast Cancer Stemness and Metastasis via TET-Family-Dependent Chromatin Remodeling. Cell 2013, 154, 311–324. [Google Scholar] [CrossRef] [Green Version]
- Huang, L.; Wang, X.; Wen, C.; Yang, X.; Song, M.; Chen, J.; Wang, C.; Zhang, B.; Wang, L.; Iwamoto, A.; et al. Hsa-MiR-19a Is Associated with Lymph Metastasis and Mediates the TNF-α Induced Epithelial-to-Mesenchymal Transition in Colorectal Cancer. Sci. Rep. 2015, 5, 13350. [Google Scholar] [CrossRef] [Green Version]




| Parameter/Feature | Breast Cancer (n = 93) | Healthy Control (n = 34) | p-Value (Chi-Squared Test) | |
|---|---|---|---|---|
| Age (Mean Years ±SD) | 46 ± 10.55 | 29 ± 7.5 | ||
| ≤35 | 16 (17.20%) | 27 (79%) | 2.19 × 10−10 | |
| >35 | 77 (82%) | 7 (20%) | ||
| <25 | 12 (12.90%) | 5 (14.7%) | ||
| BMI | 25–29.9 | 35 (37.63%) | 10 (29.4%) | |
| 30–34.9 | 24 (25.81%) | 12 (35.3%) | 0.4732 | |
| 35–39.9 | 10 (10.75%) | 7(20.5%) | ||
| ≥40 | 11.83%) | - | ||
| Missing | 1 (1.08%) | - | ||
| Histology | *IDC | 84 (90.32%) | ||
| *ILC | 8 (8.62%) | |||
| Metaplastic | 1 (1.08%) | |||
| 2 | 28 (30.11%) | |||
| 3 | 58 (62.37%) | |||
| Missing | 7 (7.53%) | |||
| Subtype | TNBC | 36 (38.71%) | ||
| Luminal A | 16 (17.20%) | |||
| Luminal B | 41 (44.09%) | |||
| ER status | positive | 55 (59.14%) | ||
| negative | 38 (40.86%) | |||
| PR status | positive | 54 (58.06%) | ||
| negative | 39 (41.94%) | |||
| HER2 | positive | 16 (17.2%) | ||
| negative | 77 (82.8%) | |||
| Tumor size | ≤2.0 cm | 19 (20.43%) | ||
| 2.1 cm–5.0 cm | 38 (40.86%) | |||
| >5.0 cm | 11 (11.83%) | |||
| Missing | 25 (26.88%) | |||
| Metastasis status | M0 | 70 (75.27%) | ||
| M1 | 21 (22.58%) | |||
| Mx | 2 (2.15%) | |||
| Lymph node | positive | 54 (58.06%) | ||
| negative | 34 (36.56%) | |||
| Missing | 5 (5.38%) | |||
| Ki67 | ≤15 | 20 (21.51%) | ||
| >15 | 73 (78.5%) | |||
| KEGG IDs | KEGG Terms | Top Hits | p-Value | Adjusted p-Value (BH) |
|---|---|---|---|---|
| hsa04151 | PI3K-Akt signaling pathway | 47 | 0 | 0 |
| hsa04110 | Cell cycle | 23 | 0 | 0 |
| hsa01521 | EGFR tyrosine kinase inhibitor resistance | 21 | 0 | 0 |
| hsa04218 | Cellular senescence | 31 | 0 | 0 |
| hsa04140 | Autophagy | 24 | 0 | 0 |
| hsa04917 | Prolactin signaling pathway | 17 | 0 | 0 |
| hsa04115 | p53 signaling pathway | 18 | 0 | 0 |
| hsa04066 | HIF-1 signaling pathway | 20 | 0.0001 | 0.0006 |
| hsa04668 | TNF signaling pathway | 20 | 0.0001 | 0.0006 |
| hsa01522 | Endocrine resistance | 18 | 0.0002 | 0.001 |
| hsa04210 | Apoptosis | 22 | 0.0002 | 0.001 |
| hsa04014 | Ras signaling pathway | 31 | 0.0003 | 0.0015 |
| hsa05235 | PD-L1 expression and PD-1 checkpoint pathway in cancer | 16 | 0.0005 | 0.0023 |
| hsa04010 | MAPK signaling pathway | 36 | 0.0005 | 0.0023 |
| hsa04068 | FoxO signaling pathway | 20 | 0.0008 | 0.0033 |
| hsa01524 | Platinum drug resistance | 13 | 0.0018 | 0.0064 |
| hsa04137 | Mitophagy | 12 | 0.002 | 0.0069 |
| hsa04012 | ErbB signaling pathway | 14 | 0.0023 | 0.0078 |
| hsa04370 | VEGF signaling pathway | 11 | 0.0029 | 0.0097 |
| hsa04150 | mTOR signaling pathway | 18 | 0.0162 | 0.0352 |
| hsa04310 | Wnt signaling pathway | 18 | 0.021 | 0.0426 |
| hsa04630 | JAK-STAT signaling pathway | 21 | 0.0034 | 0.0112 |
| hsa04350 | TGF-beta signaling pathway | 14 | 0.0052 | 0.0154 |
| hsa04152 | AMPK signaling pathway | 16 | 0.0076 | 0.0206 |
| hsa04390 | Hippo signaling pathway | 18 | 0.018 | 0.0387 |
| hsa04662 | B cell receptor signaling | 11 | 0.0229 | 0.0446 |
| hsa04660 | T cell receptor signaling | 13 | 0.0229 | 0.0446 |
| GO-BP IDs | GO-BP Terms | Top Terms/Hits | p-Value | Adjusted p-Value (BH) |
|---|---|---|---|---|
| GO:0007049 | Cell cycle | 36 | 0 | 0 |
| GO:0043066 | Negative regulation of apoptotic process | 56 | 0 | 0 |
| GO:0006974 | Cellular response to DNA damage stimulus | 33 | 0 | 0 |
| GO:0051301 | Cell division | 40 | 0.0001 | 0.007 |
| GO:0001934 | Positive regulation of protein phosphorylation | 23 | 0.0002 | 0.0123 |
| GO:0071456 | Cellular response to hypoxia | 19 | 0.0003 | 0.0147 |
| GO:0016567 | Protein ubiquitination | 47 | 0.0003 | 0.0147 |
| GO:0070317 | Negative regulation of G0 -G1 transition | 10 | 0.0004 | 0.0179 |
| GO:0030177 | Positive regulation of Wnt signaling pathway | 9 | 0.0007 | 0.0264 |
| GO:0000082 | G1-S transition of mitotic cell cycle | 16 | 0.0007 | 0.0264 |
| GO:0019221 | Cytokine mediated signaling pathway | 32 | 0.0009 | 0.0276 |
| GO:0035019 | Somatic stem cell population_ maintenance | 12 | 0.0009 | 0.0276 |
| GO:0048147 | Negative regulation of fibroblast proliferation | 8 | 0.0008 | 0.0276 |
| GO:0006470 | Protein de-phosphorylation | 20 | 0.001 | 0.0289 |
| GO:0010628 | Positive regulation of gene expression | 40 | 0.0011 | 0.03 |
| GO:0006606 | Protein import into nucleus | 13 | 0.0015 | 0.0351 |
| GO:0016055 | Wnt signaling pathway | 23 | 0.0015 | 0.0351 |
| GO:0042149 | Cellular response to glucose starvation | 9 | 0.0019 | 0.0389 |
| GO:0016579 | Protein de-ubiquitination | 29 | 0.0021 | 0.0397 |
| GO:0031647 | Regulation of protein stability | 13 | 0.0022 | 0.04 |
| GO:0045787 | Positive regulation of cell cycle | 8 | 0.0023 | 0.0403 |
| GO:0001933 | Negative regulation of protein phosphorylation | 12 | 0.0026 | 0.044 |
| GO:0032467 | Positive regulation of cytokinesis | 8 | 0.0041 | 0.0516 |
| GO:0000079 | Regulation of cyclin-dependent protein serine-threonine kinase activity | 10 | 0.0035 | 0.0516 |
| GO:0071560 | Cellular response to transforming growth factor beta stimulus | 10 | 0.0039 | 0.0516 |
| GO:0050821 | Protein stabilization | 21 | 0.004 | 0.0516 |
| GO:0001836 | Release of cytochrome c from mitochondria | 6 | 0.0039 | 0.0516 |
| GO:0014068 | Positive regulation of PI3K signaling | 13 | 0.0038 | 0.0516 |
| GO:0044772 | Mitotic cell cycle phase transition | 6 | 0.0047 | 0.0537 |
| GO:0010629 | Negative regulation of gene expression | 25 | 0.0046 | 0.0537 |
| GO:1902895 | Positive regulation of pri-miRNA transcription by RNA polymerase II | 7 | 0.0045 | 0.0537 |
| GO:0045737 | Positive regulation of cyclin-dependent protein serine-threonine kinase activity | 7 | 0.0045 | 0.0537 |
| GO:0006366 | Transcription by RNA polymerase II | 28 | 0.005 | 0.0558 |
| GO:0071230 | Cellular response to amino acid stimulus | 9 | 0.0058 | 0.0565 |
| GO:0014065 | PI3K signaling | 7 | 0.0061 | 0.0565 |
| GO:0061418 | Regulation of transcription from RNA polymerase II promoter in response to hypoxia | 11 | 0.0058 | 0.0565 |
| GO:0010507 | Negative regulation of autophagy | 9 | 0.0052 | 0.0565 |
| GO:0007265 | Ras protein signal transduction | 11 | 0.0063 | 0.0573 |
| GO:0050680 | Negative regulation of epithelial cell proliferation | 10 | 0.0065 | 0.058 |
| TNBC Drugs | miRNAs ID | Target Genes |
|---|---|---|
| 5-fluoroucil | miR-19a;miR-19b;miR-25;miR-199a-3p; miR-22;miR-93 | NFKB1; BCL2; PTEN; MSH2 |
| fluorouracil | miR-19a;miR-19b;miR-25;miR-199a-3p; miR-22;miR-93 | ABCB1;GSTT1;ABCC4;NFKB1;UGT1A1;RRM2;ABCG2;ERBB2;IGF1;IGF2;IGFBP3;TP53;BCL2;CDKN1A;ABCC3;SMUG1;TDG;MBD4;ABCC5;UPP1;UPP2;PTEN;UCK2;CLCN6;WDR7;SLC35A2;APC;RUNX3;FXYD3;FDXR;DUT;DHFR;UMPS;MTHFR;DPYD;TPMT;UPB1;FASLG;ERCC1;NOS3;GNAS;CES2;TK1;XRCC3;NT5C;GSTM1;CYP2A6;SLC19A1;KLC3;UNG |
| gefitinib | miR-19a;miR-19b;miR-25;miR-199a-3p; miR-22;miR-210;miR-93 | ABCB1;CYP3A4;PTGS2;UGT1A1;CYP1A1;ABCG2;CCND1;ABL1;APAF1;IL15;KIT;PDGFRB;ERBB2;EGF;ERBB3;IL8;IL8RA;GAB1;MET;EMP1;CYP2C9;AKT1;FUS;EGFR |
| gemcitabine | miR-19a;miR-19b;miR-25;miR-199a-3p; miR-22;miR-210;miR-93 | ABCB1;ABCC4;RRM2;ABCG2;ERBB2;BCL2;CDKN1A;SP1;PRKCA;PRKCE;XRCC5;ABCC3;ERBB3;PARP1;AICDA;ABCC5;PTEN;TOP2A;HPRT1;NT5C2;MKI67;EPC2;CLU;POLE;GPM6A;IQGAP2;TGM3;VAV3;BCL2L1;DCK;CDKN1B;ATP7B;ERCC1;POLS;USF2;DCTD;SLC28A1;AKT1;POLA2;PRKCB1;CDKN2A;SLC29A2;IGFBP1;USF1;VEGFA;SLC28A2;CMPK1;EGFR;BAX |
| bevacizumab | miR-19a;miR-19b;miR-25;miR-199a-3p; miR-22;miR-210 | ABCB1;UGT1A1;IGF1;IGF2;IGFBP3;KDR;HIF1A;VHL;DPYD;FCGR2A;FCGR3A;GSTM1 |
| alkylating agents | miR-19a;miR-19b;miR-25;miR-199a-3p; miR-22;miR-210;miR-93 | MDM2;MTHFR |
| capecitabine | miR-19a;miR-19b;miR-25;miR-199a-3p; miR-22;miR-210;miR-93 | GSTT1;PTGS2;UGT1A1;RRM2;FRAP1;UPP1;UPP2;ERCC6;GSTT1;PTGS2;UGT1A1;RRM2;FRAP1;UPP1;UPP2;ERCC6;MTHFR;CYP2C9;UGT1A1;FRAP1;DPYD;GSTA1;UPB1;ERCC1;MTHFR;UGT1A1;DCTD;FRAP1;DPYD;CES2;TK1;GSTM1;MTHFR;CYP2C9;UGT1A1;DCTD;DPYD;GSTT1;MTHFR;FRAP1;GSTT1;PTGS2;VEGFA;MTHFR;RRM2;FRAP1;DPYD;CES2;GSTA1;UPP1;UPP2;APEX2;RAD54B |
| carboplatin | miR-19a;miR-19b;miR-25;miR-199a-3p; miR-22;miR-210;miR-93 | CYP3A4;SLC22A2;MTHFR;ABCG2;ATP7A;DPYD;TP53;CDKN1A;CAMTA1;CYP1B1;MAPT;ABCB1;UGT1A1;CYP2C8;ABCC1;GSTM1;SLC19A1 |
| cisplatin | miR-19a;miR-19b;miR-25;miR-199a-3p; miR-22;miR-210;miR-199a;miR-93 | GSTT1;VEGFA;DHFR;SLC22A2;ABCC4;NQO1;MDM2;RRM2;ABCG2;ATP7A;GSTM4;GCLC;GCLM;GPX6;GNAS;ABL1;APAF1;FUS;KIT;PDGFRB;FRAP1;AKT1;MCL1;ERBB2;EGFR;DPYD;TPMT;XPC;CDKN2A;TP53;BCL2;BCL2L1;XIAP;DCK;CDKN1A;RB1;ERBB3;GSTA1;ABCC5;SLC29A2;PTEN;TOP2A;LRP2;SLC31A1;HPRT1;NT5C2;ATP7B;BAX;BID;SUMO1;CD3EAP;ATM;DNAJC15;MKI67;GSTM3;GJA1;BRCA2;EPHA2;XRCC2;TWIST1;ATP8B4;CDKN2D;EBF3;FAM57A;FCHSD1;IRF2BP2;LRRC32;MYO5B;NBEAL2;PARD6B;PGM1;PQLC3;SHMT2;SLC6A8;SORBS2;STK17A;CDK6;ABCB1;SOCS3;TOP2B;CYP2E1;UGT1A1;GPX7;IL15;XRCC5;ABCC3;PPP1R13L;HOXB9;CSF1;UMPS;SLCO1B1;GSTA4;TOP1;GATM;ARVCF;ERCC1;BAK1;DDIT4;NEK2;PFKFB4;ABCC1;GPX2;UBE2I;GALNTL4;XPA;GSTM1;GPX1;GPX3;GSTM2;GSTM5;ALDH7A1;NUF2;TMEM37;IGFBP1;CD44 |
| cetuximab | miR-19a;miR-19b;miR-25;miR-199a-3p; miR-22;miR-210;miR-93 | PTGS2;VEGFA;CCND1;EGFR;KRAS;FCGR3A;IL8;HBEGF;EGF;IL8RA;FCGR2A |
| cyclophosphamide | miR-19a;miR-19b;miR-25;miR-199a-3p; miR-22;miR-210;miR-93 | CYP3A4;GSTT1;MTHFR;DRD2;CYP1A2;ABCG2;NR1I2;ERBB2;SOD2;TP53;BCL2;CDKN1A;GSTA1;CYP1B1;CD3EAP;GSTM3;WDR7;ABCB1;VDR;CYP2E1;UGT1A1;PPP1R13L;CYP2B6;CYP2C9;CYP2C8;NR1I3;ERCC1;NOS3;GSTM1;CYP2A6 |
| dexamethasone | miR-19a;miR-19b;miR-25;miR-199a-3p; miR-22;miR-210;miR-93 | ABCB1;CYP3A4;GSTT1;PTGS2;TNF;VDR;CREBBP;EP300;AGT;CYP2E1;UGT1A1;ADRB2;CORIN;NR1I2;PIK3CA;TGFBR2;PDPK1;NR3C1;GNB1;MAP2K3;MAPK14;MYD88;TLR2;PIK3R1;IL1A;BDKRB2;IL8;IL3;SMARCD1;MAP4K4;TGFBR1;CAV1;SMAD3;SMAD4;ACTB;ARID1A;NF1;SMARCC1;CYP2B6;MTHFR;CYP1A2;CYP2C9;CYP2C8;DUSP1;TPMT;GTF2A1;GTF2E1;POLR2A;IKBKG;IL13;NOS3;GNAS;MAP2K6;SMARCE1;MAPK11;GTF2B;SMARCA4;SMARCC2;GSTM1;CYP2A6;AKT1;NPPA;MAP3K7;SLC19A1;TGFB3;IL5;IL6;IL10;CYP3A43 |
| docetaxel | miR-19a;miR-19b;miR-25;miR-199a-3p; miR-22;miR-210;miR-93 | ABCB1;CYP3A4;CYP2E1;ABCG2;ATP7A;SLC10A2;SPG7;PPARD;TNFAIP2;APAF1;NR1I2;ERBB2;IGF2;KRAS;PIK3CA;TGFBR2;TGFBR3;XRCC4;CYP2F1;EGF;TP53;BCL2;CDKN1A;ABCC5;PTEN;PLK1;CYP1B1;MAPT;IGFBP2;WDR7;BRCA2;CYP2B6;MTHFR;CYP2C9;CYP2C8;CHST3;GSTA4;DPYD;TPMT;CYP2C18;BCL2L1;RPN2;CDKN1B;ATP7B;MFAP4;ABCC1;RPL13;TMEM43;GSTM1;CYP2A6;GSTM5;IGF2AS;ABCC6;IGFBP1;SLCO1B3;NAT2;EGFR;XPC |
| doxorubicin | miR-19a;miR-19b;miR-25;miR-199a-3p; miR-22;miR-210;miR-93 | MTHFR;HRH1;NFKB1;NQO1;XDH;CAT;NOS1;CYCS;ABCG2;NR1I2;AKT1;ERBB2;SETD4;TP53;BCL2;RALBP1;HIF1A;GSTA1;CASP3;ABCC5;PTEN;TOP2A;FOXO3;CYP1B1;WDR7;MET;ERBB4;SLC19A3;ABCB1;CBR1;TOP2B;PLK1;PPP2R4;MMP1;CYP2C8;SOD1;PIM1;BAK1;OXTR;CYBA;NOS3;ABCC1;MTOR;GSTM1;AKR1A1;GPX1;SCN5A;CD44 |
| epirubicin | miR-19a;miR-19b;miR-25;miR-199a-3p; miR-22;miR-210;miR-93 | ABCB1;ERBB2;SOD2;TOP2A;NQO1;ABCC1 |
| everolimus | miR-19a;miR-19b;miR-25;miR-199a-3p; miR-22;miR-210;miR-93 | FRAP1;MKI67 |
| fulvestrant | miR-19a;miR-19b;miR-25;miR-199a-3p; miR-22;miR-93 | ESR1;ERBB2;ADORA1 |
| letrozole | miR-19a;miR-19b;miR-25;miR-199a-3p; miR-22;miR-210;miR-93 | CCND1;COLEC12;CTSK;DKK3;EGR1;GPNMB;KIAA0101;PHLDA2;CYP19A1;COL3A1;DCN;DUSP1;IRS1;SFRP4;CCNB1;MMP2;ZWINT;CYR61;HMGB2;MLF1IP;NUSAP1;SERPINA3;GEM;TPBG |
| metformin | miR-19a;miR-19b;miR-25;miR-199a-3p; miR-22;miR-210;miR-93 | ABCB1;SLC22A3;ABCG2;CCND1;ERBB2;SREBF1;RPS6KB1;CDKN1A;PRKAA1;PRKAA2;PRKAB2;STK11;SLC22A2;CYP2C9;CDKN1B;PRKAB1;PRKAG2;NDUFA1;NDUFS1;NDUFS4;SLC47A2 |
| methotrexate | miR-19a;miR-19b;miR-25;miR-199a-3p; miR-22;miR-210;miR-93 | ABCB1;CYP3A4;GSTT1;TNF;SLCO1A2;ABCC4;VDR;UGT1A1;ADRB2;SLCO4C1;ABCG2;ERBB2;MTRR;TP53;ITGB2;RALBP1;PTPRC;SPP1;NR3C1;CDKN1A;TERF1;ABCC3;CREB1;IL8RB;MTR;RFC1;IL8RA;ITGAL;ADA;HPRT1;NP;UCK2;ADORA2A;GART;ARID5B;SLC16A7;ELMO1;CLCN6;GJA1;MLL;HHEX;FTH1;GCH1;ABLIM1;ACP2;ANKRD12;AP2B1;ARF4;ARHGAP5;ARL4C;ATF1;ATRN;CA3;CLK1;CNOT8;CP;CTSL1;CUL4B;DPYSL2;DSG1;EFNB2;ETV5;FRK;FZD6;GOLGA2;GPR137B;H3F3B;HIC2;IAPP;IL1R1;JRKL;KIAA0143;KIAA1467;KIF3A;MAN2B2;MPHOSPH9;MYH15;NMT1;PARG;PCDH9;PLS1;POLE;PPP1CB;PRDM2;PRKY;PTPRG;RAB31;RAPH1;RBBP8;SACS;SAP18;SMCHD1;SS18;ST18;STK24;STK38;TGIF;TXK;WIF1;CD97;CHST1;IGFBP4;TMEM45A;DHFR;CYP2B6;MTHFR;ADORA1;XDH;SLCO1B1;DPYD;TPMT;JUN;TLR4;RB1;MSH3;SLC22A9;ITGAX;NT5E;PPAT;FAM3C;FGF9;MTHFD2;CRYZ;CSH2;GALNT7;GDF11;GZMM;HAT1;IL1RL1;MRPL33;MYLK;NUP98;PRKCQ;RDX;TESK1;TOB2;YY1;NOS3;ABCC1;TK1;MAX;ADORA3;ADORA2B;AP1S2;IL1RN;NRXN2;RAB5C;RCC1;SSX1;GSTM1;G6PD;ABCC11;SLC22A11;SLC19A1;GBF1;MPO;ITPA;AMT;ADRA1D;CST7;DEFA4;GBE1;GCHFR;GDF10;LCN2;PTK7;FOLR1;PECAM1;PTS;SLC22A8;SLCO1B3;NAT2;APP;AHCY;EIF4A1;BAX;E2F1;BYSL;FZD2;GNG10;PF4V1;PRG1;RNASE6;TCEB3 |
| paclitaxel | miR-19a;miR-19b;miR-25;miR-199a-3p; miR-22;miR-210;miR-93 | ABCB1;CYP3A4;ABCG2;NR1I2;ERBB2;TP53;BCL2;CDKN1A;CASP3;FOXO3;CYP1B1;AKT2;MAPT;WDR7;APC;CYP2B6;MTHFR;CYP1A2;CYP2C8;SLCO1B1;DPYD;BAK1;ABCC1;PHB;GSTM1;CYP2A6;VEGFA;SLCO1B3; CD44 |
| Platinum/platinum compounds | miR-19a;miR-19b;miR-25;miR-199a-3p; miR-22;miR-210;miR-93 | ATP7A;ATP7B;SLC22A3;SLC22A2 |
| taxol | miR-19a;miR-19b;miR-25;miR-199a-3p; miR-22;miR-210;miR-93 | PTEN;CSF1 |
| topotecan | miR-19a;miR-19b;miR-25;miR-199a-3p; miR-22;miR-210;miR-93 | ABCB1;CYP3A4;ABCG2;NR1I2;ERBB2;TP53;BCL2;PTEN;WDR7;MTHFR |
| vincristine | miR-19a;miR-19b;miR-25;miR-199a-3p; miR-22;miR-210;miR-93 | ABCB1;CYP3A4;GSTT1;VDR;UGT1A1;ABCG2;NR1I2;BCL2;NR3C1;AAK1;MTHFR;TPMT;DCK;GSTA1;ABCC1;FLT3;NPM1;GSTM1;SLC19A1;ABCC6;XIAP |
| olaparib | miR-93;miR-19a;miR-19b;miR-199a-3p; miR-22 | PTGS2;PARP1;KDR;CCR4 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Qattan, A.; Al-Tweigeri, T.; Alkhayal, W.; Suleman, K.; Tulbah, A.; Amer, S. Clinical Identification of Dysregulated Circulating microRNAs and Their Implication in Drug Response in Triple Negative Breast Cancer (TNBC) by Target Gene Network and Meta-Analysis. Genes 2021, 12, 549. https://doi.org/10.3390/genes12040549
Qattan A, Al-Tweigeri T, Alkhayal W, Suleman K, Tulbah A, Amer S. Clinical Identification of Dysregulated Circulating microRNAs and Their Implication in Drug Response in Triple Negative Breast Cancer (TNBC) by Target Gene Network and Meta-Analysis. Genes. 2021; 12(4):549. https://doi.org/10.3390/genes12040549
Chicago/Turabian StyleQattan, Amal, Taher Al-Tweigeri, Wafa Alkhayal, Kausar Suleman, Asma Tulbah, and Suad Amer. 2021. "Clinical Identification of Dysregulated Circulating microRNAs and Their Implication in Drug Response in Triple Negative Breast Cancer (TNBC) by Target Gene Network and Meta-Analysis" Genes 12, no. 4: 549. https://doi.org/10.3390/genes12040549
APA StyleQattan, A., Al-Tweigeri, T., Alkhayal, W., Suleman, K., Tulbah, A., & Amer, S. (2021). Clinical Identification of Dysregulated Circulating microRNAs and Their Implication in Drug Response in Triple Negative Breast Cancer (TNBC) by Target Gene Network and Meta-Analysis. Genes, 12(4), 549. https://doi.org/10.3390/genes12040549