Karyotype Variability and Inter-Population Genomic Differences in Freshwater Ostracods (Crustacea) Showing Geographical Parthenogenesis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Specimens
2.2. Chromosome Preparations
2.3. DNA Extraction and Whole Genomic Probe Preparation
2.4. Comparative Genomic Hybridization
2.5. Fluorescence In Situ Hybridization with 28S rDNA
2.6. Microscopy
3. Results
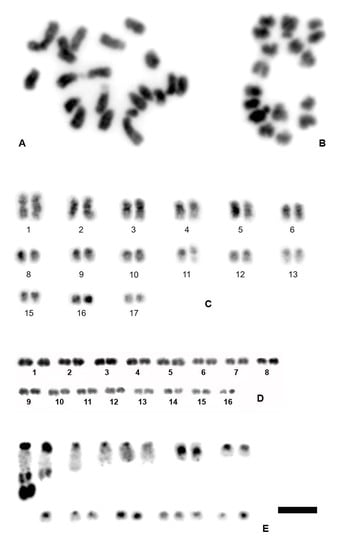
3.1. Chromosome Numbers and Karyotypes in Different E. virens Populations and Other Species
3.2. Molecular Cytogenetic Studies of E. virens Genomes by Inter-Population CGH
3.2.1. Comparative Genomic Hybridization of Almodovar and Rivalazzetto Populations
3.2.2. Comparative Genomic Hybridization of Greece and Morocco Populations
3.2.3. Interspecies Comparative Genomic Hybridization Comparison of Eucypris virens and Bradleystrandesia fuscata
3.3. Fluorescence In Situ Hybridization with 28S rDNA
3.4. Meiosis in Asexually Reproducing Ostracods
4. Discussion
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Pandian, T.J. Reproduction and development in aquatic invertebrates. In Reproduction and Development in Crustacea; CRC Press Taylor & Francis Group: Boca Raton, FL, USA, 2016; Volume 1. [Google Scholar]
- Adolfsson, S.; Michalakis, Y.; Paczesniak, D.; Bode, S.N.S.; Butlin, R.K.; Lamatsch, D.K.; Martins, M.J.F.; Schmit, O.; Vandekerkhove, J.; Jokela, J. Evaluation of elevated ploidy and asexual reproduction as alternative explanations for geographic parthenogenesis in Eucypris virens ostracods: Polyploidy and geographic lineage distribution. Evolution 2010, 64, 986–997. [Google Scholar] [CrossRef] [PubMed]
- Bode, S.N.S.; Adolfsson, S.; Lamatsch, D.K.; Martins, M.J.F.; Schmit, O.; Vandekerkhove, J.; Mezquita, F.; Namiotko, T.; Rossetti, G.; Schön, I.; et al. Exceptional cryptic diversity and multiple origins of parthenogenesis in a freshwater ostracod. Mol. Phylogenet. Evol. 2010, 54, 542–552. [Google Scholar] [CrossRef] [PubMed]
- Hartmann, G. Dr. H.G. Bronns Klassen und Ordnungen des Tierreichs; Band 5 (Arthropoda), Abt. I (Crustacea), Buch 2, Teil IV; C.F. Winter; Akademische Verlagsgesellschaft: Leipzig, Germany, 1966; pp. 1–1067. (In German) [Google Scholar]
- Horne, D.J.; Baltanas, A.; Paris, G. Geographical distribution of reproductive modes in living non-marine ostracods. In Sex and Parthenogenesis: Evolutionary Ecology of Reproductive Mode in Non-Marine Ostracods; Backhuys: Leiden, The Netherlands, 1998; pp. 77–99. [Google Scholar]
- Bell, G. The Masterpiece of Nature: The Evolution and Genetics of Sexuality; University of California Press: Berkeley, CA, USA, 1982. [Google Scholar]
- Butlin, R.K.; Schön, I.; Martens, K. Asexual reproduction in nonmarine ostracods. Heredity 1998, 81, 473–480. [Google Scholar] [CrossRef]
- Meisch, C. Freshwater Ostracoda of Western and Central Europe; Springer: Heidelberg, Germany, 2000. [Google Scholar]
- Rossetti, G.; Martens, K. Redescription and morphological variability of Darwinula stevensoni (Brady & Robertson, 1870) (Crustacea, Ostracoda). Biologie 1996, 66, 73–92. [Google Scholar]
- Vandel, A. La parthénogénèse geographique. Contribution à ľétude biologique et cytologique de la parthénogénèse naturelle. Bull. Biol. Fr. Belg. 1928, 62, 164–281. [Google Scholar]
- Stebbins, G.L. Variation and evolution in plants. Science 1950, 112, 764–766. [Google Scholar]
- Vandel, A. La parthénogénèse geographique. IV Polyploidie et Distribution Géographique. Bull. Biol. Fr. Belg. 1940, 74, 94–100. (In French) [Google Scholar]
- Stenberg, P.; Lundmark, M.; Knutelski, S.; Saura, A. Evolution of clonality and polyploidy in a weevil system. Mol. Biol. Evol. 2003, 20, 1626–1632. [Google Scholar] [CrossRef] [PubMed]
- Kearney, M. Hybridization, glaciation and geographical parthenogenesis. Trends. Ecol. Evol. 2005, 20, 495–502. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.J.; Ha, M.; Soltis, D. Polyploidy: Genome obesity and its consequences. New Phytol. 2007, 174, 717–720. [Google Scholar] [CrossRef] [PubMed]
- Beaton, M.J.; Hebert, P.D.N. Geographical parthenogenesis and polyploidy in Daphnia pulex. Am. Nat. 1988, 132, 837–845. [Google Scholar] [CrossRef]
- Havel, J.E.; Hebert, P.D.N. Apomictic parthenogenesis and genotypic diversity in Cypridopsis vidua (Ostracoda: Cyprididae). Heredity 1989, 62, 383–392. [Google Scholar] [CrossRef]
- Havel, J.E.; Hebert, P.D.N. Clonal diversity in parthenogenetic ostracods. In Ostracoda in the Earth and Life Sciences; A. A. Balkema: Roterdam, The Netherlands, 1993; pp. 353–368. [Google Scholar]
- Rossi, V.; Schön, I.; Butlin, R.K.; Menozzi, P. Clonal genetic diversity. In Sex and Parthenogenesis: Evolutionary Ecology of Reproductive Modes in Non-Marine Ostracods; Backhuys Publ.: Leiden, The Netherlands, 1998; pp. 257–274. [Google Scholar]
- Blackman, R.L.; Spence, J.M.; Normark, B.B. High diversity of structurally heterozygous karyotypes and rDNA arrays in parthenogenetic aphids of the genus Trama (Aphididae: Lachninae). Heredity 2000, 84, 254–260. [Google Scholar] [CrossRef] [PubMed]
- Gassner, M.; Dejaco, T.; Schönswetter, P.; Marec, F.; Arthofer, W.; Schlick-Steiner, B.C.; Steiner, F.M. Extensive variation in chromosome number and genome size in sexual and parthenogenetic species of the jumping-bristletail genus Machilis (Archaeognatha). Ecol. Evol. 2014, 4, 4093–4105. [Google Scholar] [CrossRef] [PubMed]
- Liehr, T.; Buleu, O.; Karamysheva, T.; Bugrov, A.; Rubtsov, N. New Insights into Phasmatodea Chromosomes. Genes 2017, 8, 327. [Google Scholar] [CrossRef] [PubMed]
- Majtánová, Z.; Choleva, L.; Symonová, R.; Ráb, P.; Kotusz, J.; Pekárik, L.; Janko, K. Asexual Reproduction Does Not Apparently Increase the Rate of Chromosomal Evolution: Karyotype Stability in Diploid and Triploid Clonal Hybrid Fish (Cobitis, Cypriniformes, Teleostei). PLoS ONE 2016, 11, e0146872. [Google Scholar] [CrossRef] [PubMed]
- Schneider, M.C.; Cella, D.M. Karyotype Conservation in 2 Populations of the Parthenogenetic Scorpion Tityus serrulatus (Buthidae): rDNA and Its Associated Heterochromatin Are Concentrated on Only One Chromosome. J. Heredity 2010, 101, 491–496. [Google Scholar] [CrossRef] [PubMed]
- Neiman, M.; Sharbel, T.F.; Schwander, T. Genetic causes of transitions from sexual reproduction to asexuality in plants and animals. J. Evol. Biol. 2014, 27, 1346–1359. [Google Scholar] [CrossRef] [PubMed]
- Tetart, J. Étude des garnitures chromosomiques de quelques Ostracodes d’eau douce. Bull. Soc. Zoo. Fr. 1967, 92, 167–179. (In French) [Google Scholar]
- Tetart, J. Les garnitures chromosomiques des ostracodes d’eau douce. Trav. Lab. Hydrobiol. 1978, 69–70, 113–140. (In French) [Google Scholar]
- Turgeon, J.; Hebert, P.D.N. Genetic characterization of breeding systems, ploidy levels and species boundaries in Cypricercus (Ostracoda). Heredity 1995, 75, 561–570. [Google Scholar] [CrossRef]
- Kubanc, S.N. Chromosome numbers of certain ostracod populations in the vicinity of Istanbul. J. Fisheries Aquatic Sci. 2002, 13, 53–64. [Google Scholar]
- Kubanc, S.N.; Gűlen, D. Chromosome numbers of three Ostracoda (Crustacea) species from Küçükçekmece Lake. J. Biol. 2002, 65, 95–107. [Google Scholar]
- Bianchi-Bullini, A.P.; Bullini, L. Osservazioni sul corredo cromosomico Eucypris virens e di Cypris bispinosa (Crustacea Ostracoda). Accademia Nazionale dei Lincei—Rend. Classe di Scienze Fisiche, Matematiche e Naturali 1972, LII, 59–63. (In Italian) [Google Scholar]
- Gregory, T.R. Animal Genome Size Database. Available online: http://genomesize.com (accessed on 1 December 2017).
- Jeffery, N.W.; Ellis, E.A.; Oakley, T.H.; Gregory, T.R. The Genome Sizes of Ostracod Crustaceans Correlate with Body Size and Evolutionary History, but not Environment. J. Hered. 2017, 108, 701–706. [Google Scholar] [CrossRef] [PubMed]
- Adolfsson, S.; Lamatsch, D.K.; Paczesniak, D.; Michalakis, Y.; Martens, K.; Schön, I.; Butlin, R.K.; Jokela, J. Mitochondrial cluster-specific genome size variability among sexual and asexual lineages of the ostracod Eucypris virens species group. Joannea Geol. Paläont. 2011, 11, 9–12. [Google Scholar]
- Seidl, M.F.; Thomma, B.P. Sex or no sex: Evolutionary adaptation occurs regardless. BioEssays 2014, 36, 335–345. [Google Scholar] [CrossRef] [PubMed]
- Martins, M.J.F.; Vandekerkhove, J.; Namiotko, T. Environmental stability and the distribution of the sexes: insights from life history experiments with the geographic parthenogen Eucypris virens (Crustacea: Ostracoda). Oikos 2008, 117, 829–836. [Google Scholar] [CrossRef]
- Schmit, O.; Bode, S.N.S.; Camacho, A.; Horne, D.J.; Lamatsch, D.K.; Martens, K.; Martins, M.J.F.; Namiotko, T.; Rossetti, G.; Rueda-Sevilla, J.; et al. Linking present environment and the segregation of reproductive modes (geographical parthenogenesis) in Eucypris virens (Crustacea: Ostracoda). J. Biogeogr. 2013, 40, 2396–2408. [Google Scholar] [CrossRef]
- Vandekerkhove, J.; Martens, K.; Rossetti, G.; Mesquita-Joanes, F.; Namiotko, T. Experimental assessment of the fecundity of Eucypris virens (Ostracoda, Crustacea) under natural sex ratios. Freshw. Biol. 2007, 52, 1058–1064. [Google Scholar] [CrossRef]
- Petkovski, T.; Fuhrmann, R. Die Gattung Bradleystrandesia Broodbakker (Crustacea, Ostracoda) in Europa. Mauritiana 2009, 20, 587–600. [Google Scholar]
- Symonová, R. Microanatomy and cytogenetics of non-marine ostracods-an insight into evolutionary biology of their reproductive modes. Ph.D. Thesis, Charles University, Prague, Czech Republic, 2009. [Google Scholar]
- Dietz, R. Zahl und Verhalten der Chromosomen einiger Ostracoden. Zeitschrift für Naturforschung B 1955, 10, 92–95. (In German) [Google Scholar] [CrossRef]
- Dhar, M.K.; Kaul, S. FISH reveals somatic association of nucleolus organizing regions in Plantago ovata. Curr. Sci. 2004, 87, 1336–1337. [Google Scholar]
- Scali, V.; Coluccia, E.; Deidda, F.; Lobina, C.; Deiana, A.M.; Salvadori, S. Co-localization of ribosomal and telomeric sequences in Leptynia (Insecta: Phasmatodea). Ital. J. Zool. 2016, 83, 285–290. [Google Scholar] [CrossRef]
- Bugrov, A.G.; Karamysheva, T.V.; Rubtsov, D.N.; Andreenkova, O.V.; Rubtsov, N.B. Comparative FISH analysis of distribution of B chromosome repetitive DNA in A and B chromosomes in two subspecies of Podisma sapporensis (Orthoptera, Acrididae). Cytogenet. Genome Res. 2004, 106, 284–288. [Google Scholar] [CrossRef] [PubMed]
- Bugrov, A.G.; Karamysheva, T.V.; Perepelov, E.A.; Elisaphenko, E.A.; Rubtsov, D.N.; Warchałowska-Śliwa, E.; Tatsuta, H.; Rubtsov, N.B. DNA content of the B chromosomes in grasshopper Podisma kanoi Storozh. (Orthoptera, Acrididae). Chromosome Res. 2007, 15, 315–326. [Google Scholar] [CrossRef] [PubMed]
- LéCher, P.; Defaye, D.; Noel, P. Chromosomes and nuclear DNA of Crustacea. Invertebr. Repr. Dev. 1995, 27, 85–114. [Google Scholar] [CrossRef]
- Cabrero, J.; Lopez-Leon, M.D.; Bakkali, M.; Camacho, J.P. Common origin of B chromosome variants in the grasshopper Eyprepocnemis plorans. Heredity 1999, 83, 435–439. [Google Scholar] [CrossRef] [PubMed]
- Camacho, J.P.M.; Sharbel, T.F.; Beukeboom, L.W. B-chromosome evolution. Biol. Sci. 2000, 355, 163–178. [Google Scholar] [CrossRef] [PubMed]
- Bakkali, M.; Cabrero, J.; López-León, M.D.; Perfectti, F.; Camacho, J.P.M. The B chromosome polymorphism of the grasshopper Eyprepocnemis plorans in North Africa. I. B variants and frequency. Heredity 1999, 83, 428–434. [Google Scholar] [CrossRef] [PubMed]
- Bakkali, M.; Camacho, J.P.M. The B chromosome polymorphism of the grasshopper Eyprepocnemis plorans in North Africa: III. Mutation rate of B chromosomes. Heredity 2004, 92, 428–433. [Google Scholar] [CrossRef] [PubMed]
- Coluccia, E.; Cannas, R.; Cau, A.; Deiana, A.M.; Salvadori, S. B chromosomes in Crustacea decapoda. Cytogenet. Genome Res. 2004, 106, 215–221. [Google Scholar] [CrossRef] [PubMed]







| Species | Country | Population | Sex/Asex | 2n/3n | Geographic Coordinates |
|---|---|---|---|---|---|
| E. virens | Italy | Rivalazzetto (It051) | Asex | 3n | N44°49′07″ E10°08′20″ |
| E. virens | Italy | Sicily (It064) | Sex | 2n | N37°53′27″ E14°22′50″ |
| E. virens | Spain | Almodovar | Asex | 2n-3n | N38°44′15″ W04°09′11″ |
| E. virens | Spain | Valencia, Triops Pond | Asex | 2n | N40°25′06″ W00°04′24″ |
| E. virens | Spain | Laguna Caracuel | Sex | 2n | N38°49’33″ W04°04’01″ |
| E. virens | Morocco | Sidi Chiker | Sex | 2n | N31°48′42″ W08°32′42″ |
| E. virens | Spain | San Carlos, Bull Pond | Mixed | 2n-3n | N39°54′22″ W06°03′38″ |
| E. virens | Greece | Corfu, Skripera | Sex | 2n | N39°41′52″ E19°47′08″ |
| E. virens | France | Normandy, Calvados | Asex | 3n | N49°11′22″ E00°07′13′′ |
| C. pubera | Germany | Gauting, oxbow | Asex | 2n | N48°02.455′ E11°22.393′ |
| H. incongruens | Germany | Gauting, oxbow | Asex | 2n | N48°02.455′ E11°22.393′ |
| H. incongruens | Germany | Martinsried, Munich | Asex | 2n | N48°06′42.5″ E11°27′47.5″ |
| H. incongruens | Italy | Sicily, Zingaro | Asex | 2n | N37°53′27″ 14°22′50″ |
| B. fuscata | Germany | Gauting, oxbow | Asex | 2n | N48°02.455′ E11°22.393′ |
| C. marginata | Germany | Gauting, oxbox | Sex | 2n | N48°02.455′ E11°22.393′ |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Symonová, R.; Vrbová, I.; Lamatsch, D.K.; Paar, J.; Matzke-Karasz, R.; Schmit, O.; Martens, K.; Müller, S. Karyotype Variability and Inter-Population Genomic Differences in Freshwater Ostracods (Crustacea) Showing Geographical Parthenogenesis. Genes 2018, 9, 150. https://doi.org/10.3390/genes9030150
Symonová R, Vrbová I, Lamatsch DK, Paar J, Matzke-Karasz R, Schmit O, Martens K, Müller S. Karyotype Variability and Inter-Population Genomic Differences in Freshwater Ostracods (Crustacea) Showing Geographical Parthenogenesis. Genes. 2018; 9(3):150. https://doi.org/10.3390/genes9030150
Chicago/Turabian StyleSymonová, Radka, Iva Vrbová, Dunja K. Lamatsch, Jürgen Paar, Renate Matzke-Karasz, Olivier Schmit, Koen Martens, and Stefan Müller. 2018. "Karyotype Variability and Inter-Population Genomic Differences in Freshwater Ostracods (Crustacea) Showing Geographical Parthenogenesis" Genes 9, no. 3: 150. https://doi.org/10.3390/genes9030150
APA StyleSymonová, R., Vrbová, I., Lamatsch, D. K., Paar, J., Matzke-Karasz, R., Schmit, O., Martens, K., & Müller, S. (2018). Karyotype Variability and Inter-Population Genomic Differences in Freshwater Ostracods (Crustacea) Showing Geographical Parthenogenesis. Genes, 9(3), 150. https://doi.org/10.3390/genes9030150